edit: Results are current as of Dec 4, 2018 13:00 PST.
Background
K-mers have many uses in bioinformatics, and for this reason it would be useful to know the most RAM-efficient and fastest way to work with them programmatically. There have been questions covering what canonical k-mers are, how much RAM k-mer storage theoretically takes, but we have not yet looked at the best data structure to store and access k-mers and associated values with.
Question
What data structure in C++ simultaneously allows the most compact k-mer storage, a property, and the fastest lookup time? For this question I choose C++ for speed, ease-of-implementation, and access to lower-level language features if desired. Answers in other languages are acceptable, too.
Setup
- For benchmarking:
- I propose to use a standard
fastafile for everyone to use. This program,generate-fasta.cpp, generates two million sequences ranging in length between 29 and 300, with a peak of sequences around length 60. - Let's use
k=29for the analysis, but ignore implementations that require knowledge of the k-mer size before implementation. Doing so will make the resulting data structure more amenable to downstream users who may need other sizesk. - Let's just store the most recent read that the k-mer appeared in as the property to retrieve during k-mer lookup. In most applications it is important to attach some value to each k-mer such as a taxon, its count in a dataset, et cetera.
- If possible, use the string parser in the code below for consistency between answers.
- The algorithm should use canonical k-mers. That is, a k-mer and its reverse complement are considered to be the same k-mer.
- I propose to use a standard
Here is generate-fasta.cpp. I used the command g++ generate_fasta.cpp -o generate_fasta to compile and the command ./generate_fasta > my.fasta to run it:
return 0;
//generate a fasta file to count k-mers
#include <iostream>
#include <random>
char gen_base(int q){
if (q <= 30){
return 'A';
} else if ((q > 30) && (q <=60) ){
return 'T';
} else if ((q > 60) && (q <=80) ){
return 'C';
} else if (q > 80){
return 'G';
}
return 'N';
}
int main() {
unsigned seed = 1;
std::default_random_engine generator (seed);
std::poisson_distribution<int> poisson (59);
std::geometric_distribution<int> geo (0.05);
std::uniform_int_distribution<int> uniform (1,100);
int printval;
int i=0;
while(i<2000000){
if (i % 2 == 0){
printval = poisson(generator);
} else {
printval = geo(generator) + 29;
}
if (printval >= 29){
std::cout << '>' << i << '\n';
//std::cout << printval << '\n';
for (int j = 0; j < printval; j++){
std::cout << gen_base(uniform(generator));
}
std::cout << '\n';
i++;
}
}
return 0;
}
Example
One naive implementation is to add both the observed k-mer and its reverse complement as separate k-mers. This is obviously not space efficient but should have fast lookup. This file is called make_struct_lookup.cpp. I used the following command to compile on my Apple laptop (OS X): clang++ -std=c++11 -stdlib=libc++ -Wno-c++98-compat make_struct_lookup.cpp -o msl.
#include <fstream>
#include <string>
#include <map>
#include <iostream>
#include <chrono>
//build the structure. measure how much RAM it consumes.
//then measure how long it takes to lookup in the data structure
#define k 29
std::string rc(std::string seq){
std::string rc;
for (int i = seq.length()-1; i>=0; i--){
if (seq[i] == 'A'){
rc.push_back('T');
} else if (seq[i] == 'C'){
rc.push_back('G');
} else if (seq[i] == 'G'){
rc.push_back('C');
} else if (seq[i] == 'T'){
rc.push_back('A');
}
}
return rc;
}
int main(int argc, char* argv[]){
using namespace std::chrono;
//initialize the data structure
std::string thisline;
std::map<std::string, int> kmer_map;
std::string header;
std::string seq;
//open the fasta file
std::ifstream inFile;
inFile.open(argv[1]);
//construct the kmer-lookup structure
int i = 0;
high_resolution_clock::time_point t1 = high_resolution_clock::now();
while (getline(inFile,thisline)){
if (thisline[0] == '>'){
header = thisline.substr(1,thisline.size());
//std::cout << header << '\n';
} else {
seq = thisline;
//now add the kmers
for (int j=0; j< thisline.size() - k + 1; j++){
kmer_map[seq.substr(j,j+k)] = stoi(header);
kmer_map[rc(seq.substr(j,j+k))] = stoi(header);
}
i++;
}
}
std::cout << " -finished " << i << " seqs.\n";
inFile.close();
high_resolution_clock::time_point t2 = high_resolution_clock::now();
duration<double> time_span = duration_cast<duration<double>>(t2 - t1);
std::cout << time_span.count() << " seconds to load the array." << '\n';
//now lookup the kmers
inFile.open(argv[1]);
t1 = high_resolution_clock::now();
int lookup;
while (getline(inFile,thisline)){
if (thisline[0] != '>'){
seq = thisline;
//now lookup the kmers
for (int j=0; j< thisline.size() - k + 1; j++){
lookup = kmer_map[seq.substr(j,j+k)];
}
}
}
std::cout << " - looked at " << i << " seqs.\n";
inFile.close();
t2 = high_resolution_clock::now();
time_span = duration_cast<duration<double>>(t2 - t1);
std::cout << time_span.count() << " seconds to lookup the kmers." << '\n';
}
Example output
I ran the above program with the following command to log peak RAM usage. The amount of time the lookup of all k-mers in two million sequences is reported by the program. /usr/bin/time -l ./msl my.fasta.
The output was:
-finished 2000000 seqs.
562.864 seconds to load the array.
- looked at 2000000 seqs.
368.734 seconds to lookup the k-mers.
1046.94 real 942.38 user 78.96 sys
11680514048 maximum resident set size
So, the program used 11680514048 bytes = 11.68GB of RAM and it took 368.734 seconds to lookup the k-mers in two million fasta files.
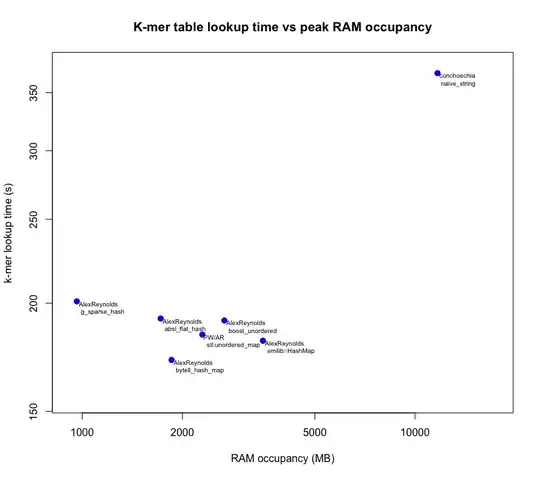
Results
Below is a plot of the results from each user's answers.
std::unordered_mapinstead ofstd::mapas a simple improvement (hopefully) – Peter Menzel Nov 04 '18 at 09:50generate-fasta.cppdoes not compile. I also don’t understand its logic; why the multiple random distributions? What are you modelling? Thegen_basefunction confuses me. – Konrad Rudolph Nov 20 '18 at 12:57gen_basefunction uses a random int between 1-100 as input to select a base. The ranges in the function give it some AT bias, The Poisson distribution makes a bunch of sequences around 59 bp long, but the geometric distribution gives it a long tail of sequence lengths. In this case I was modeling a distribution of some sequences I need to look up kmers for. – conchoecia Nov 21 '18 at 00:47