I haven't done any signal processing in a while, but I'm completely stymied by the instability and bias in my implementation of a normalized least mean squares filter.
I checked the usual things:
- Am I actually near the convergence lower bound (variance of AWGN in test signal)? No.
- Is my step size below $\frac{2}{trace[\mathbf{R}]}$ (where $\mathbf{R}$ is just an outer product of the signal used as instantaneous estimate)? Yes.
- Is my dot product of windowed past samples using the convention of increasing delay indices ("reverse" order)? Yes.
- Am I using the previous signal index when I compute predicted value, not the current one? Yes.
I narrowed down my issue by implementing a simple 1-tap filter. My reference is the algorithm on page 324 of Haykin's Adaptive Filter Theory, and I'm using the test conditions on page 280 of the same:
- Input: first-order AR process, $a = -0.99$ with noise variance $\sigma_u^2 = 0.93627$
- Trial count: 100 trials
- Signal length: 500 samples
Error
According to Haykin, I should be below $10^{-1}$ MMSE after less than 100 samples. That's not what I'm seeing, but I'm convinced it has to be something basic.
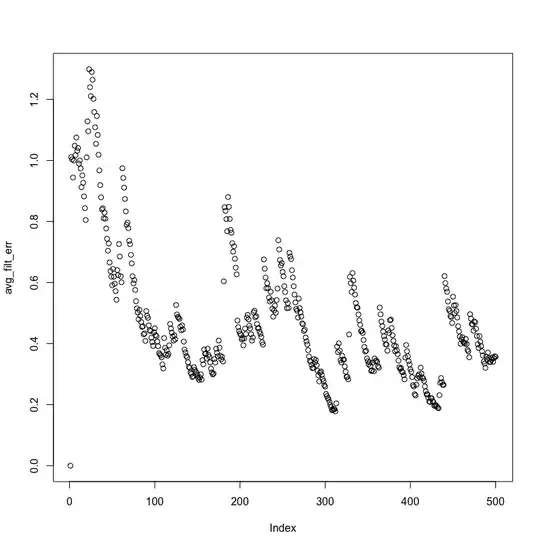
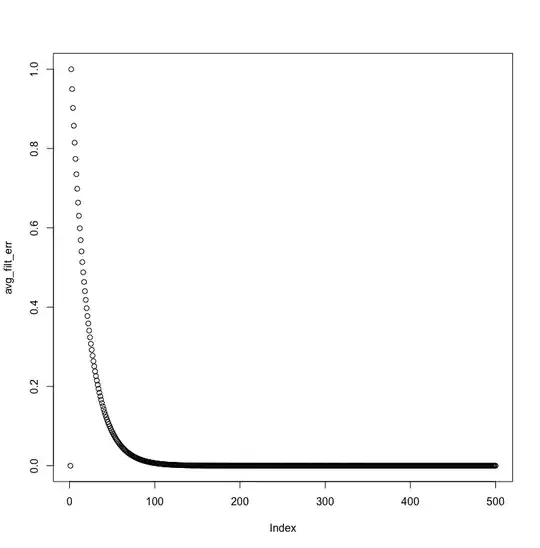
Here are MMSE and average absolute filter coefficient error, visualized:
The MMSE doesn't decrease exponentially, and the filter coefficient never comes closer than a distance of 2 from the actual AR process value.
And in fact, the MMSE is still much farther than the noise variance near the end of the signal:
[10.260998119102881, 16.060967587632792, 41.72963567568045, 49.232814770283696, 50.56247908944245, 84.29056731939356, 99.74962522053087, 74.94509263319398, 61.55708377260941, 43.79321013379245, 47.68084407515727, 51.84551183735604, 23.977556992678352, 23.691841969230886, 15.292586007185905, 12.710425182987175, 14.689863202271942, 15.03357374836029, 16.305144820997945, 21.770075252230768, 16.501486407946132, 21.62300216817481, 15.461623713499273, 26.354991476964795, 28.001121132610393, 21.632068009342056, 15.812522324768985, 15.522990356558456, 53.24514663841896, 9.36613637386399, 52.24856602614516, 98.50333720012102, 251.59105479441695, 193.85612892514402, 124.53671103656986, 159.16459102635278, 82.2544192379602, 50.25510472853462, 67.03420998875504, 9.59758344071307, 9.770861267272064, 21.975670899135086, 28.84699022842896, 29.070465314026674, 15.05395348320232, 20.20596174124456, 30.609319675498526, 14.396998393433874, 17.654986587869196, 9.664719322902583]
Filter code
To produce the above results, I tried implementing the simplest possible 1-tap NLMS filter I could, which is given below.
extern crate rand;
use std::f64::EPSILON;
use rand::distributions::{IndependentSample, Normal};
extern crate gnuplot;
use gnuplot::{Figure, Caption, Color};
fn main() {
let trial_count = 100;
let signal_len = 500;
let noise_std_dev = 0.96761;
// AR process coefficient [n-1]
let process = [-0.99];
let mut rand_gen = rand::OsRng::new().unwrap();
let mut mmse: Vec<f64> = vec![0.0; signal_len];
let mut avg_filt_err: Vec<f64> = vec![0.0; signal_len];
let noise_dist = Normal::new(0.0, noise_std_dev);
for _ in 0..trial_count {
let mut sig_vec: Vec<f64> = Vec::with_capacity(signal_len);
// Zero padding to start noise process
sig_vec.push(0.0);
for n in 1..signal_len {
let noise = noise_dist.ind_sample(&mut rand_gen);
let p1 = if n > 0 {
-process[0]*sig_vec[n-1]
} else {
0.0
};
sig_vec.push(noise + p1);
}
let step_size = 0.05;
let mut pred_vec: Vec<f64> = Vec::with_capacity(signal_len);
let mut filt_coef: f64 = 0.0;
// Zero padding to start filter process
pred_vec.push(0.0);
for sig_ind in 1..signal_len {
let pred = filt_coef * sig_vec[sig_ind - 1];
pred_vec.push(pred);
let err = sig_vec[sig_ind] - pred;
mmse[sig_ind] += err.powi(2) / trial_count as f64;
filt_coef += step_size * sig_vec[sig_ind - 1] * err /
(sig_vec[sig_ind - 1].powi(2) + EPSILON);
avg_filt_err[sig_ind] += (process[0] - filt_coef).abs() / trial_count as f64;
}
}
let x: Vec<usize> = (0..mmse.len()).collect();
let mut fg = Figure::new();
fg.axes2d()
.lines(&x, &mmse, &[Caption("MMSE"), Color("black")]);
let mut fg2 = Figure::new();
fg2.axes2d()
.lines(&x, &avg_filt_err, &[Caption("Average filter error"), Color("red")]);
fg.show();
fg2.show();
}








slice_windowfunction was missing. If you replace that code, you should be able to replicate my results (Octave 4.0.3) – bright-star Apr 29 '17 at 19:55