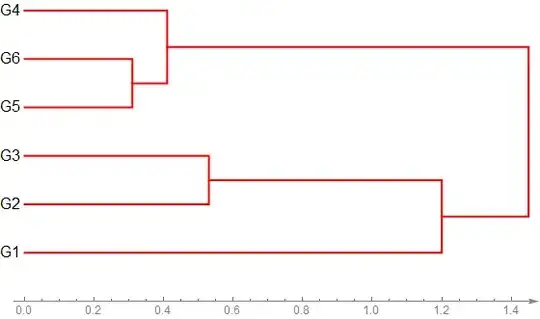
I have a distance matrix data, and want to visualize it as below:
The current codes I am using is:
data={{0., 1.2, 1.73, 3.57, 3.22, 3.17}, {1.2, 0., 0.53, 2.37, 2.05,
1.97}, {1.73, 0.53, 0., 1.85, 1.54, 1.45}, {3.57, 2.37, 1.85, 0.,
0.42, 0.41}, {3.22, 2.05, 1.54, 0.42, 0., 0.31}, {3.17, 1.97, 1.45,
0.41, 0.31, 0.}};
DendrogramPlot[DirectAgglomerate[data,Style[#, 16] & /@ {"G1", "G2", "G3", "G4", "G5", "G6"},Linkage -> "Single"], LeafLabels -> Automatic,Orientation -> Right,
PlotStyle -> {Red, Thick}, Axes -> {True, False},AxesOrigin -> {-1, 0}, ImageSize -> 560, AxesStyle -> {{Directive[Gray, 12], Arrowheads[.02]}, Automatic},AspectRatio -> 1/2]
which results:
The cluster distances information can be easily obtained by:
DirectAgglomerate[data,Style[#,16]&/@{"G1","G2","G3","G4","G5","G6"},Linkage->"Single"]
which is:
Cluster[Cluster[G1,Cluster[G2,G3,0.53,1,1],1.2,1,2],Cluster[Cluster[G5,G6,0.31,1,1],G4,0.41,2,1],1.45,3,3]
how can I label the cluster distances into the DendrogramPlot automatically?
PS: I noticed there is already a similar post here: , but the answer does not label the distances as text something like the Python implementation here.



Orientation->Right. For other orientations, similar rules can be constructed. – kglr May 15 '16 at 13:40line : Line[{_, {x_, y1_}, {x_, y2_}, _}] :> Sequence[line, {Black, Text[Style[NumberForm[x, {3, 2}], 12], {x, (y1 + y2)/2}, {1.25, 0}]}]This also would have to be modified or generalized to handle a different Orientation – Bob Hanlon May 15 '16 at 14:08