In this question I am asking about the different results I get between NIntegrate-ing a function of two variables vs. "doing it myself" with my own implementation of Simpson's rule. For various "complicated" test functions, e.g.
testfunc = Sin[#1]^2 Cos[#2]^2 Exp[-#1^2/100 - #2^2/144]/(Abs[#1 - #2] + 0.01) &
the NIntegrate of this agrees very well with my home-cooked integrator. But in the application I'm working on, the results differ significantly. I need help to understand this. What follows might seem complicated/long but I've included everything so that a) you can just copy-paste it and it will work, and b) I cannot seem reproduce the problem for a simpler integrand.
The reason I have tried by own home-cooked integrator is that I want to integrate as below for many different values of en, and so I am willing to sacrifice a few per cent accuracy for speed. However, I seem to be sacrificing way too much, see below.
I have the following basic definitions:
Clear[vCI]
vCI[q1_, q2_, qs_, kappa_, d_] :=
2 Pi/(kappa (Sqrt[(q1^2 + q2^2)/2] + qs)) Exp[-Sqrt[(q1^2 + q2^2)/
2]*d]
vCI[q1_, q2_, qs_, kappa_, 0] :=
2 Pi/(kappa (Sqrt[(q1^2 + q2^2)/2] + qs))
Clear[spinor]
spinor[q1_, q2_, xi_, m_, vF_] :=
With[{alpha = vF*q2, beta = q1^2/(2 m)},
With[{en = xi Sqrt[alpha^2 + beta^2]},
{beta + xi (en + alpha), en - alpha + xi beta}
]
]
Clear[overlap]
overlap[spinor1_, spinor2_] :=
If[Chop[spinor1] == {0, 0} || Chop[spinor2] == {0, 0},
0,
(spinor1.spinor2)^2/((spinor1.spinor1) (spinor2.spinor2))
]
Clear[energy]
energy[q1_, q2_, xi_, m_,
vF_] :=
xi Sqrt[(vF*q2)^2 + (q1^2/(2*m))^2]
Now I define
m = 0.05;
vF = 5.;
qscreen = 0.01;
kappa = 1.;
d = 0.;
eta = 0.01;
ni = 1;
q1 = 0.5;
q2 = 0.5;
xi = 1;
en = energy[q1, q2, xi, m, vF];
and importantly
SetAttributes[integrand, Listable];
integrand[q1p_, q2p_] := Re[ni*vCI[q1 - q1p, q2 - q2p, qscreen, kappa, d]^2*
overlap[spinor[q1, q2, xi, m, vF],
spinor[q1p, q2p, 1, m, vF]]/(en -
energy[q1p, q2p, 1, m, vF] + I eta)]
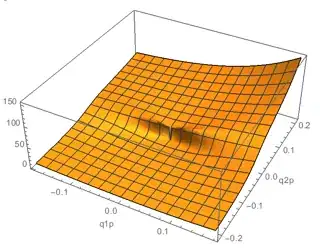
A plot of the integrand in the interesting region:
Plot3D[
integrand[q1p, q2p], {q1p, -0.18, 0.18}, {q2p, -0.21, 0.21}
, PlotPoints -> 50, AxesLabel -> Automatic
]
Looks fairly tame, except perhaps around q2p == 0. I'll talk more about this later.
We can integrate it:
NIntegrate[
integrand[q1p, q2p], {q1p, -0.18, 0.18}, {q2p, -0.21, 0.21}]
(* 5.46624 + 0. I *)
NB: Not really sure why there is a complex part here (even though it's zero) because Re should give a purely real result even for approximate complex input.
Ok, now to the home-cooked integrator using Simpson's rule:
simpsonIntegrate[f_][q1pmin_, q1pmax_, q2pmin_, q2pmax_, numPoints_] :=
Block[{a = q1pmin, b = q1pmax, c = q2pmin, d = q2pmax, n = numPoints,
h, k, range, q1even, q1odd, q2even, q2odd},
h = (b - a)/(2 n);
k = (d - c)/(2 n);
range = Range[2*n - 1];
q1even = (a + h*range)[[2 ;; ;; 2]];
q1odd = (a + h*range)[[1 ;; ;; 2]];
q2even = (c + k*range)[[2 ;; ;; 2]];
q2odd = (c + k*range)[[1 ;; ;; 2]];
h*k/9.*(
f[a, c] + f[a, d] + f[b, c] + f[b, d] +
4.*Total[
f[a, q2odd] + f[b, q2odd] + f[q1odd, c] + f[q1odd, d]] +
2.*Total[
f[a, q2even] + f[b, q2even] + f[q1even, c] + f[q1even, d]] +
16.*Total[Map[Total[f[#, q2odd]] &, q1odd]] +
8.*(Total[Map[Total[f[#, q2odd]] &, q1even]] +
Total[Map[Total[f[#, q2even]] &, q1odd]]) +
4.*Total[Map[Total[f[#, q2even]] &, q1even]]
)
]
The formula is taken from here. I'm using the listability as much as I can, which is the reason for the Map[Total[...]] stuff. Using the testing function above gives
NIntegrate[testfunc[x, y], {x, -10, 10}, {y, -12, 12}]
(* 32.1532 *)
simpsonIntegrate[testfunc][-10, 10, -12, 12, 100]
(* 32.3869 *)
and improving if I increase the last argument to simpsonIntegrate. But look:
simpsonIntegrate[integrand[#1, #2] &][-0.18, 0.18, -0.21, 0.21, 200]
(* 4.52673 *)
This is a deviation of 20% from NIntegrate! Increasing the number of gridpoints per dimension to 1000 does not help. Why this discrepancy?
I first assumed it had something to do with the line q2p == 0 as mentioned before. We can make this go away:
SetAttributes[integrand2, Listable];
integrand2[q1p_, q2p_] := integrand[q1p, q2p]*Boole[Abs[q2p] > 0.02]
Now there definitely shouldn't be a problem, right? Wrong:
NIntegrate[
integrand2[q1p, q2p], {q1p, -0.18, 0.18}, {q2p, -0.21, 0.21}]
(* 4.9732 + 0. I *)
simpsonIntegrate[integrand2[#1, #2] &][-0.18, 0.18, -0.21, 0.21, 200]
(* 4.11283 *)
Still 20% off... Why?
FINAL NOTE VERSION 2
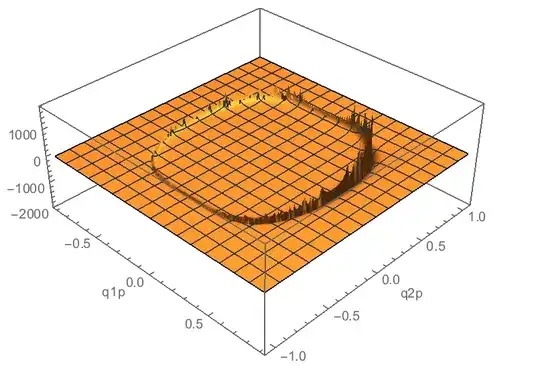
In the end I would like to increase the range of integration, but the integrand becomes nastier. I've changed qscreen to 1 here:
Plot3D[
integrand[q1p, q2p], {q1p, -0.18/0.2, 0.18/0.2}, {q2p, -0.21/0.2,
0.21/0.2}
, AxesLabel -> Automatic, PlotPoints -> 50, PlotRange -> All
]
I also need to do this integral for many different values of en, so if I can get a quick result that is consistently no more than 2-5% wrong, I would be happy with that accuracy.


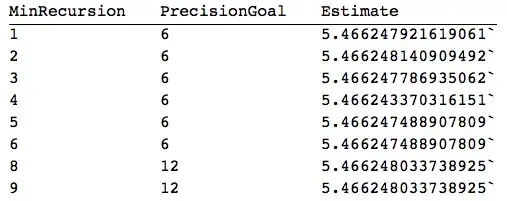

Interpolation[]withInterpolationOrder -> 2, and thenIntegrate[]the resulting function. – J. M.'s missing motivation Jun 13 '16 at 11:50integrand2shouldn't Simpson's then overestimate the result, i.e. give a small contribution when the function drops to zero instead of nothing? Also, the error should be smaller and smaller still for finer mesh, right? – Marius Ladegård Meyer Jun 13 '16 at 11:56Interpolationtrick like you suggested and it gave a very good result, 5.46775. How can that be? Is there an error in mysimpsonIntegrateafter all? – Marius Ladegård Meyer Jun 13 '16 at 13:14testfunc. The first thing I did was a simple Gaussian, runs fine. Can you check my routine on your end for some lame function, ortestfunc? Might have someClearing to do here, but can't do it right now... – Marius Ladegård Meyer Jun 13 '16 at 13:22