It has already been pointed out in this question that using the element mesh of the interpolating function greatly reduces the time it takes to compute numerical integrals over the whole domain of the function.
Can a similar optimization be used to compute and plot the marginal distributions? For example, let $\psi(x_1,x_2)$ be some computed eigenfunction, and interpret $\lvert \psi(x_1,x_2) \rvert^2$ as a probability distribution, then how can we quickly find $$ M_1(x_1) = \int \lvert \psi(x_1,x_2) \rvert^2 \ dx_2$$ $$ M_2(x_2) = \int \lvert \psi(x_1,x_2) \rvert^2 \ dx_1?$$
For the case of two quantum mechanical particles in a 1D infinite well, with an inter-particle interaction $V_{int} \sim \alpha (x_1-x_2)^2$ we can use NDEigensystem to find the lowest 3 eigenfunctions numerically.
\[Alpha] = -(1/10000);
{Evals, Efuncts} =
NDEigensystem[{-Laplacian[\[Psi][x1, x2], {x1, x2}] + (\[Alpha] (x1 - x2)^2) \[Psi][x1, x2],
DirichletCondition[\[Psi][x1, x2] == 0, True]}, \[Psi], {x1, 0, 1}, {x2, 0, 1}, 3];
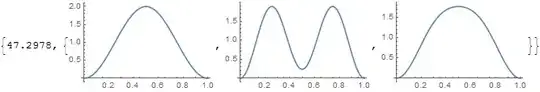
Then plot the single particle probability distributions for the particle $x_2$, what I have just called $M_2(x_2)$ above
AbsoluteTiming[ParallelTable[Plot[Quiet[
NIntegrate[Efuncts[[i]][x1, x2] Efuncts[[i]][x1, x2], {x1, 0, 1}, MaxRecursion -> 100]],
{x2, 0, 1}, PlotRange -> All], {i, 1, Length[Efuncts]}]]
Although solving the system is very fast, plotting the single particle probability distributions takes almost a minute (sometimes significantly longer, depending on the value of $\alpha$). In contrast, doing almost any integral over the entire domain of interest using the aforementioned optimization takes less than a second.
AbsoluteTiming[ParallelTable[NIntegrate[
Exp[-x1^2 - x2^2]*Efuncts[[i]][x1, x2] Efuncts[[i]][x1, x2], {x1, x2} \[Element]
Efuncts[[1]]["ElementMesh"]], {i, 1, 3}]]
{0.732146, {0.587017, 0.564539, 0.587082}}