I am trying to do several hundred iterations of these fully coupled recursion equations (but will use twenty iterations for the example).
ClearAll["Global`*"]
x1[t_] := x1[t] = (1 - m) x1[t - 1] + m x2[t - 1]
x2[t_] := x2[t] = (1 - m) x2[t - 1] + m x1[t - 1]
x1[0] := 1
x2[0] := 0
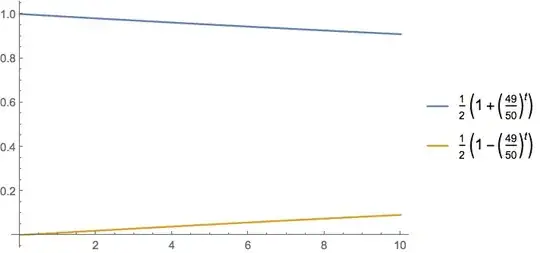
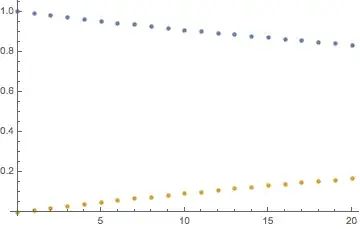
ListPlot[Table[{{t, x1[t]}, {t, x2[t]}} /. m -> 0.01, {t, 0,
20}]] // AbsoluteTiming
Memoization sped it up from 27.933541 seconds to 7.681030 seconds but I think it takes so long because it is still recursively calculating all the values from the other equation. I have read several posts about partially coupled recursion equations (e.g. Solve pair of recurrence relations, How do I use RSolve to solve a system of recurrence relations?) but ultimately, I won't be able to use RSolve because my equations will be too complicated and I cannot find a solution to speed up iterations of fully coupled recursion equations.
Is there a way to link x1[t] and x2[t] together for memoization? Or is a For loop the way to go? Thanks!
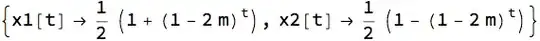
parameters = {m->0.01}but can beparameters = {m->0.01, s->0.1, alpha ->0.5}– biologyUser Sep 15 '16 at 22:00DiscretePlot[{x1[t, 0.01], x2[t, 0.01]}, {t, 0, 20}, Filling -> None]– Karsten7 Sep 16 '16 at 08:03