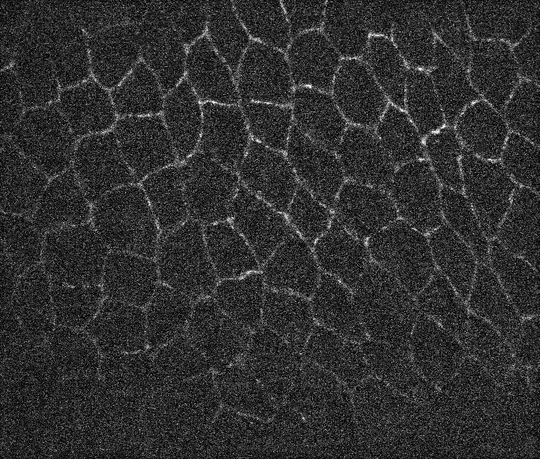
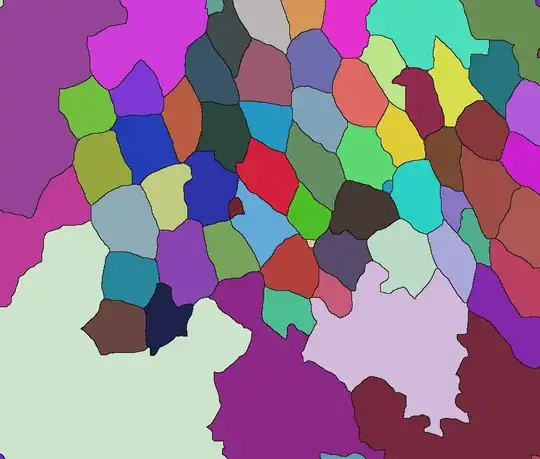
I have a microscope image of some animal tissue and wish to get the contours for all the cells that are present in the image. the cells are connected to the neighbouring cells via these contours. At the bottom of the image the signal intensity is faint but the human eye can still detect some contours.
I have tried a bunch of techniques including the use of ClusteringComponents and MorphologicalBinarize, LaplacianGaussianFilter and GradientFilter but have been unsuccessful in my approaches. The particular problem I am facing is the inability to get rid of the noisy signal (grains/granules whatever you may wish to call them) inside the contours during segmentation.
Can anyone kindly help me for my research problem. Thanks in advance.
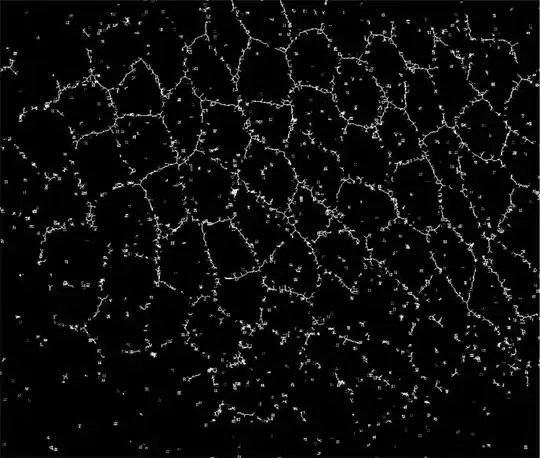
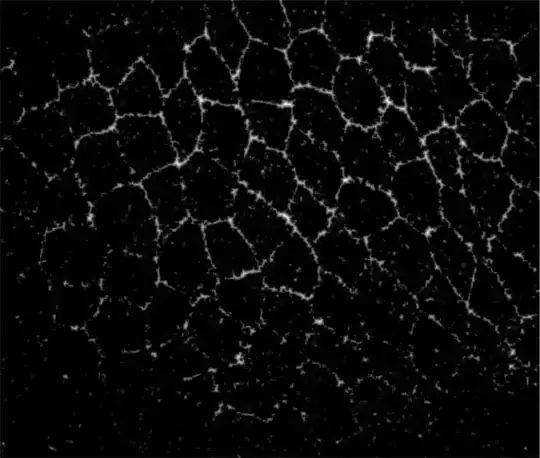
the closest i have are the following approaches but they do not prove satisfactory:
KuwaharaFilter[CommonestFilter[GaussianFilter[
Binarize[img, 0.2, Method -> "MinimumError"], 3], 3],3]
Using SkeletonTransform after KuwaharaFilter and application of other filters
KuwaharaFilter[CommonestFilter[GaussianFilter[
Binarize[img, 0.18, Method -> "MinimumError"], 3],3], 3] //
Binarize[#, 0.6] & // SkeletonTransform
Using DistanceTransform in conjunction with KuwaharaFilter and a bunch of filters
KuwaharaFilter[CommonestFilter[GaussianFilter[
Binarize[img, 0.18, Method -> "MinimumError"], 3],3], 3] //
Binarize[#, 0.55] & //DistanceTransform // ImageAdjust