I'm interested in simulating chemical reactions with perturbations. I can simulate a given reaction using NDSolve ("rxn"} with a given added noise component ("noise1"). Due to the noise, each evaluation gives a unique result. Can I output the result of each evaluation as a new column in a table? I'd like to evaluate the NDSolve i times, each using a different value of noise1[t], and then output given time points to a table. Simply iterating in the Table[] function, gives identical outputs since noise1[t_] is only evaluated once.
I've provided some simplified sample code below:
noise1[t_] := RandomReal[NormalDistribution[0, 0.2]]
totaltime = 10;
rxn = NDSolve[{
a'[t] == -a[t] + noise1[t],
a[0] == 10},
a,
{t, 0, totaltime}];
Plot[a[t] /. rxn, {t, 0, totaltime}]
output = Table[Flatten[{{t}, a[t] /. rxn}], {t, 0, totaltime}];
MatrixForm[output]
desiredoutput = {{t, "a[t]-evaluation 1", "a[t]-evaluation 2",
"a[t]-evaluation 3", "..."}
Currently, I can simulate a bunch of trajectories by exporting a csv for each evaluation, and joining the tables. I know this is the kookiest way possible, but I haven't been able to figure out a better approach.
EDIT: Thanks to @m_goldberg, I've built a functioning model. For this simple model, A -> B, and both concentrations fluctuate. Fluctuations are made by creating a noise function v[t] (see Adding noise to nonlinear control system using NDSolve, can't provide link due to reputation requirement :-/). The code below simulates n1 reactions, over a time 0 to tmax, using a noise function with n2 points, with a distribution sigma. The final data is provided as a table, as well as list plots of all trajectories. Distribution of the product concentration ([B]) is taken at the end. True thermodynamic fluctuation for a given elementary step, A -> B can be approximated by the equation below: 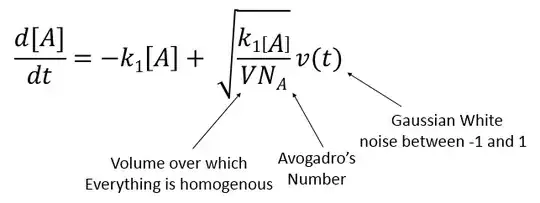
(* Fluctuation Parameters *)
odeSols[n1_, tmax_, n2_, sigma_] :=
Table[With[{v = Interpolation@Join[{{0, 0.}},
Rest@Table[{t, RandomReal@NormalDistribution[0, sigma]},
{t, 0, tmax, (tmax)/(n2 - 1)}]]},
NDSolveValue[
{a'[t] == -0.1 a[t] + 0.1 v[t], b'[t] == 0.1 a[t] + 0.1 v[t],
a[0] == 0.4, b[0] == 0},
{a, b}, {t, 0, tmax}]], n1]
result[n1_, tmax_, n2_, sigma_] :=
Module[{asol, bsol, tvals, aplotdata, bplotdata, adata, bdata, data,
header},
tvals = Range [0, tmax];
asol = odeSols[n1, tmax, n2, sigma][[All, 1]];
bsol = odeSols[n1, tmax, n2, sigma][[All, 2]];
adata = Transpose @ Join[Table[asol[[i]] /@ tvals, {i, n1}]];
bdata = Transpose @ Join[Table[bsol[[i]] /@ tvals, {i, n1}]];
data = Table[
Prepend[Flatten[Append[adata[[i]], bdata[[i]]]], tvals[[i]]], {i,
tmax}];
header =
Prepend[Flatten[
Append[Table["a[t]- " <> ToString[i], {i, n1}],
Table["b[t]- " <> ToString[i], {i, n1}]]], "t"];
aplotdata =
Table[Partition[
Riffle[Table[tvals[[i]], {i, tmax}], Transpose[adata][[j]]],
2], {j, 1, n1}];
bplotdata =
Table[Partition[
Riffle[Table[tvals[[i]], {i, tmax}], Transpose[bdata][[j]]],
2], {j, 1, n1}];
ListLinePlot[bplotdata, ImageSize -> Large,
PlotLabel -> Style["[B]", FontSize -> 36]]
ListLinePlot[aplotdata, ImageSize -> Large,
PlotLabel -> Style["[A]", FontSize -> 36]]
SmoothHistogram[Last[bdata], ImageSize -> Large,
PlotLabel -> Style["Distribution of [B] at tmax", FontSize -> 24]]
Grid[Join[{header}, data],
Frame -> All,
Alignment -> {Center, Automatic},
ItemSize -> All]]
result[30, 30, 200, 0.2]
Below are examples of the output:


