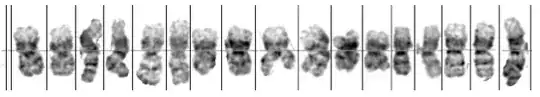
I'm aware of the solution to a similar problem by Hayashi Yoshiaki. Is there a way of extending the solution to break out the chromosomes from this example I have a mass of data in this format that needs to be converted to a training set. Note the there are holes in the chromosome that the human ignores when segmenting. Doing this manually for hundreds of images is a challenge. Thanks in advance
Asked
Active
Viewed 180 times
3
1 Answers
1
Here's the very straightforward method I suggested in my comment:
tot = Total@ImageData[Binarize[im, 0.9]];
columnsSequence = Flatten@Position[tot, x_ /; x >= 103];
columns = Round@(Mean /@ Split[columnsSequence, #2 - #1 == 1 &]);
imd = ImageData[im];
imd[[All, columns, All]] = 0;
Image[imd]
This is just drawing the lines that separate the chromosomes. From here you can split the image or do any other further manipulation. For calculating the best separating "white column" I used this trick.
Here's how to divide the image to subimages:
imd=ImageData[ims]
images = Table[
imd[[All, columns[[i]] ;; columns[[i + 1]], All]], {i,
Length[columns] - 1}]
Image /@ images
yohbs
- 7,046
- 3
- 29
- 60
-
OK, its a good start - how do I generate a list of images from this? I'll post a page of data from one example in the morning. – Boris Jul 31 '17 at 22:31
-
Hi yohbs, Ive tested your routine on other sets and find that I need to adjust the position parameter but thats OK I can automate that. Im having trouble breaking out the individual components due to my limited familiarity with array handling. I shall persevere. – Boris Aug 01 '17 at 10:28
-
-
pt1 = Drop[Partition[Riffle[columns, 0], 2], -1]; pt2 = Drop[Partition[Riffle[columns, 116], 2], 1]; components = Partition[Partition[Flatten[Transpose[{pt1, pt2}]], 2], 2]; images = ImageTrim[im, #] & /@ components – Boris Aug 01 '17 at 14:00
-
-


ImagePartitions[image, Scaled[{1/17,1}]. At this time, that approach doesn't work because the widths are different for different chromosomes unfortunately. – MarcoB Jul 31 '17 at 19:14