I have created an empirical distribution that given an array containing the points px (e.g.{px0,px1,px2,px100}) the correspondent probability is given by a liner interpolation of the tipe:
The probability density function (PDF) corresponds to the angular coefficient m of the lines, which is computed in the function below.
GeneralEmpiricalDistribution[xdata_, pt_] :=
Block[{n, tab, cdf, pdf, sz, i, index, m, x, x0, y0, xf, yf},
sz = Length[xdata];
n = sz - 2;
tab = Table[{xdata[[i]], (i - 1)/(n + 1)}, {i, 1, n + 2}];
For[i = 1, i < sz, i++,
If[Between[pt, {xdata[[i]], xdata[[i + 1]]}], index = i; Break[] ];
If[i == sz - 1, index = sz - 1]
];
{x0, y0} = tab[[index]];
{xf, yf} = tab[[index + 1]];
m = (yf - y0)/(xf - x0);
cdf = m (x - x0) + y0;
pdf = m;
{cdf, pdf}
]
Then to validate the function above, consider the data (xdata) generated from a normal distribution with mean=2, and sdev=0.4:
mean = 2;
sdev = 0.4;
xdata = Table[Quiet[NSolve[CDF[NormalDistribution[mean, sdev], x] == prob, x][[1,1,2]]], {prob, 0.00000029, 1., 0.00999971}];
n = Length[xdata];
xmax = xdata[[n]];
Plot[{GeneralEmpiricalDistribution[xdata, x][[2]],
PDF[NormalDistribution[mean, sdev], x]}, {x, 0, xmax}]
Which seems to be OK. But when i try to integrate to find the mean again the solution are very different:
NIntegrate[x GeneralEmpiricalDistribution[xdata, x][[2]], {x, 0, xmax}]
NIntegrate[x PDF[NormalDistribution[mean, sdev], x], {x, 0, xmax}]
Results:
0.0959046
1.99989
Why the integration results are different?
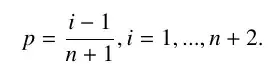

NIntegratedoesn't seem to be holding its first argument, so you're integratingGeneralEmpiricalDistribution[xdata, x]. Which is basically a flat-line. – b3m2a1 Sep 11 '17 at 20:08