Here is a modified version of my answer to How to numerically solve a 1-d time-independent Schrödinger equation? that solves this problem.
I use NDEigensystem after manually shifting the potential so its bottom is at zero energy. This way the eigenvalues are automatically sorted in ascending order. In the output, I undo that shift so the eigenvalues are given on the original energy scale which has its zero at the point where the potential becomes non-binding. You can see that the largest eigenvalue is above zero, so that all the results up to this value represent the complete list of bound states.
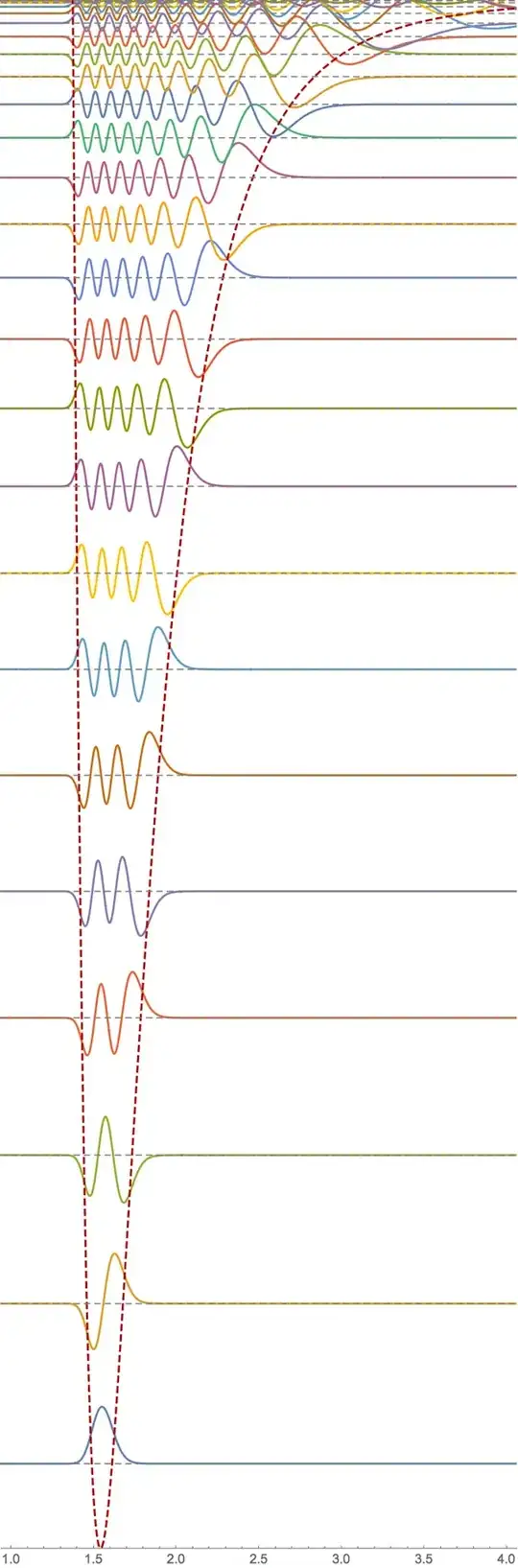
In the second half of the code, I plot the eigenfunctions, superimposed on the potential with a baseline corresponding to their eigenvalue.
V[x_] := 102*(4343/x^12 - 650/x^6) + 33/x^2
n = 25;
cutoffDistance = 10;
shift = -FindMinValue[V[x], x];
{ev, ef} =
NDEigensystem[{shift f[x] + V[x] f[x] - 1/2 f''[x],
DirichletCondition[f[x] == 0, True]}, f, {x, 0, cutoffDistance},
n, Method -> {"SpatialDiscretization" -> {"FiniteElement", \
{"MeshOptions" -> {"MaxCellMeasure" -> 0.001}}},
"Eigensystem" -> {"Arnoldi", "MaxIterations" -> 10000}}];
evShifted = ev - shift
(*
==> {-2332.04, -2076.97, -1840.3, -1621.45, -1419.88, -1235.01, \
-1066.23, -912.955, -774.545, -650.364, -539.751, -442.027, -356.495, \
-282.438, -219.119, -165.779, -121.639, -85.8998, -57.7403, -36.3193, \
-20.776, -10.2314, -3.79112, -0.553967, 0.450964}
*)
With[{amplitudes = Table[30, n]},
Show[Plot[
Evaluate[
Table[evShifted[[i]] + amplitudes[[i]] ef[[i]][x], {i, n}]], {x,
0, cutoffDistance},
PlotRange -> {-shift, Max[evShifted] + Max[amplitudes]},
Epilog -> {Gray, Dashed,
Table[Line[{{0, evShifted[[i]]}, {cutoffDistance,
evShifted[[i]]}}], {i, n}]}, AspectRatio -> 3],
Plot[V[x], {x, 0.0001, cutoffDistance},
PlotStyle -> Directive[Dashed, Darker@Red], PlotRange -> All],
ImageSize -> 400
]]

To see the plots more clearly, you would have to zoom in on the graphics, or restrict the PlotRange, as I do here (also increased number of PlotPoints):
With[{amplitudes = Table[30, n]},
Show[Plot[
Evaluate[
Table[evShifted[[i]] + amplitudes[[i]] ef[[i]][x], {i, n}]], {x,
0, cutoffDistance}, PlotPoints -> 100,
PlotRange -> {-shift, Max[evShifted] + Max[amplitudes]},
Epilog -> {Gray, Dashed,
Table[Line[{{0, evShifted[[i]]}, {cutoffDistance,
evShifted[[i]]}}], {i, n}]}, AspectRatio -> 3],
Plot[V[x], {x, 0.0001, cutoffDistance},
PlotStyle -> Directive[Dashed, Darker@Red], PlotRange -> All],
PlotRange -> {{1, 4}, {-shift, 0}}, AxesOrigin -> {0, -shift},
ImageSize -> 400
]]