How can I visualize the normal mode vibrations of a molecule in Mathematica? Something like the following GIF:
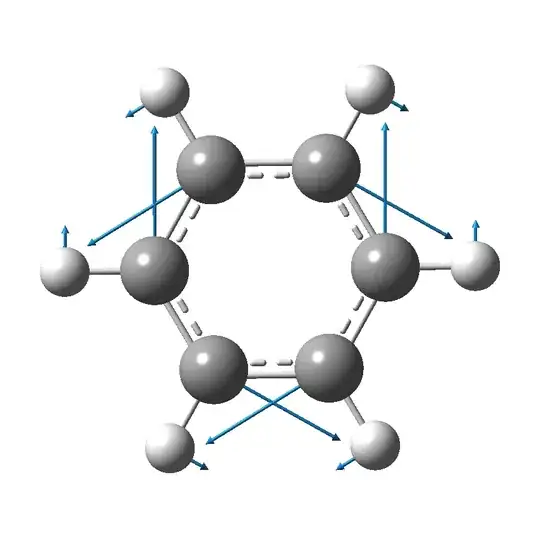 (from here)
(from here)
I have the XYZ file of the benzene molecule and the normal mode coordinates.
How can I visualize the normal mode vibrations of a molecule in Mathematica? Something like the following GIF:
 (from here)
(from here)
I have the XYZ file of the benzene molecule and the normal mode coordinates.
Using a few undocumented functions that have been discussed here on the stack exchange before,
{atoms, coords} = Import[
"https://www.dropbox.com/s/q3rwpedngv5wbqi/ben.xyz?dl=1",
{"XYZ", {"VertexTypes", "VertexCoordinates"}}
];
mode = Most @ Import[
"https://www.dropbox.com/s/3pjxams7ndr0l7h/19.mode?dl=1", "Table"
];
mode = 100 * mode;
bonds = Graphics`MoleculePlotDump`InferBonds[atoms, coords, 40, 25];
range = Range[-0.5, 0.5, 0.025];
range = Join[range, Reverse @ range];
plot[v_] := ImportExport`MoleculePlot3D[
{
"VertexCoordinates" -> (coords + v * mode),
"VertexTypes" -> atoms, "EdgeRules" -> bonds
}
];
ListAnimate @ Map[plot, range]
When importing the mode file, it returns an empty list as the last element so I need to use Most to make it a matrix. Then I multiplied it by 100 to convert to picometers. Finally I had to precompute the bonds, because as the molecule moves along the normal coordinate, the atoms could get far enough apart that the bonds wouldn't be detected by MoleculePlot3D.
Sources:
ImageSize option to the plotting function.
– Jason B.
Dec 01 '18 at 14:36
.modefile? Is there a standard file format for vibrational modes? – Jason B. Nov 30 '18 at 21:26