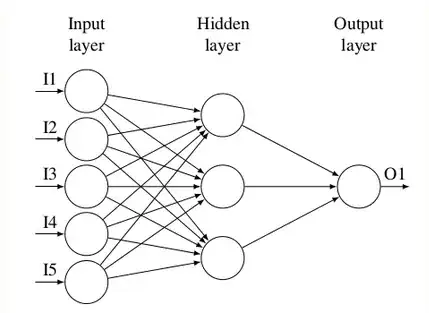
Using CompleteGraph with a list of layer sizes as the first argument and deleting undesired edges:
ClearAll[nwG]
nwG[layers : {__}, opts : OptionsPattern[Graph]] := Module[{nf = First@layers,
nl = Last@layers, cg = CompleteGraph[Flatten[{First@layers, layers, Last@layers}],
DirectedEdges -> True]},
cg = EdgeDelete[cg, {DirectedEdge[a_, b_] /;
(Subtract @@ (PropertyValue[{cg, #}, VertexCoordinates][[1]] & /@ {b, a}) > 1),
DirectedEdge[v1_, v2_] /; Or[And[v1 <= nf, v2 != nf + v1],
And[v2 >= 1 + VertexCount[cg] - nl, v1 != v2 - nl]]}];
SetProperty[cg, {PerformanceGoal -> "Quality",
VertexShapeFunction ->
{Alternatives @@ Join[Take[VertexList[cg], nf], Take[VertexList[cg], -nl]] :> None},
VertexSize -> .5, VertexStyle -> White,
EdgeStyle -> Black, EdgeLabelStyle -> 16,
VertexCoordinates -> (ReflectionTransform[{0, -1}]@ GraphEmbedding[cg]),
EdgeLabels -> {DirectedEdge[v : Alternatives @@ Range[nf], _] :>
Placed[Subscript["I", v], 1/2],
## & @@ MapIndexed[DirectedEdge[_, #] -> Placed[Subscript["O", #2[[1]]], 1/2] &,
Take[VertexList[cg], -nl]]}, opts}]]
Examples:
g0 = nwG[{5, 3, 1}, ImageSize -> Large]

SetProperty[g0, {VertexSize -> .9, VertexStyle -> {11 -> Yellow},
VertexCoordinates -> (ScalingTransform[{2, 1}]@GraphEmbedding[g0])}]

layers = {5, 2, 4, 3, 2, 1, 2, 3, 4};
nwG[layers, VertexStyle -> {23 -> Red}, ImageSize -> Large, ImagePadding -> Scaled[.03],
Epilog -> {Text["First\nLayer", {1, 4}], Text["Third\nLayer", {3, 4}],
Text["Seventh\nLayer", {7, 4}]}]