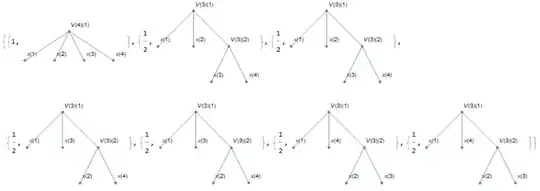
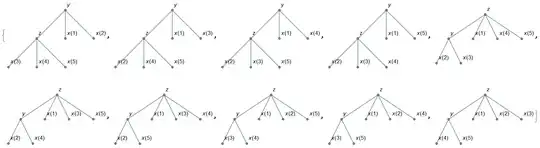
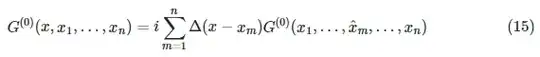Here is a dumbed down version of the algorithm presented here, based on the description provided here. Hopefully, this more simplistic (less efficient?) code might be more readable to demonstrate the concept.
We will be generating vertices with index labels on the fly, so we define a function that produces unique vertex labels when called:
ClearAll[cnt]
(* given a head, return it with a unique, automatically incremented index *)
newVar[x_] := If[ValueQ[cnt[x]], cnt[x] = cnt[x] + 1; x[cnt[x]], cnt[x] = 1; x[cnt[x]]]
Then we define the lowest level (1-valent vertex) graphs described by
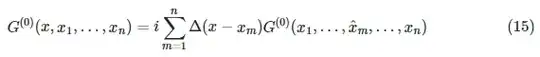
ClearAll[G]
G[0][{a_, b_}] = Ε[x[a], x[b]]; (* simple edge *)
G[_][{a_}] = 0; (* no infinitely long edges starting at a point *)
G[_][{}] = 0;(* no self-looping edges *)
(* recursively construct all graphs of 1-valent vertices *)
G[0][{a_, c__}] := Sum[Ε[x[a], x[b]] G[0][DeleteCases[{c}, b]], {b, {c}}]
where we use Ε[a,b] to denote an edge.
Next we define a function that does the higher order recursion

(* perturbative Schwinger-Dyson eq. recursion*)
G[k_][{a_, c__}] /; k > 0 := Module[{tmp},
Sum[
tmp = newVar[V[q]];
1/(q - 1)! Ε[x[a],x[tmp]] G[k + 2 - q][{Sequence @@ Table[tmp, q - 1], c}]
, {q, 3, k + 2}]
+ Sum[
Ε[x[a], x[b]] G[k][ DeleteCases[{c}, b]]
, {b, {c}}] // Expand
]
Note that instead of only 4-valent vertices as in the picture, we include all 3<=q<=k+2 valent vertices that can possibly contribute at order k, which is a simple generalization.
In principle, the above already generates the sums of products of edges. Now we just want to represent these as lists of lists of edges, and also draw them as graphs. Also, the unique vertex labels reach high numbers, which we can relabel back to low numbers in each term as follows:
(* this resets larger number head indices to minimal indices in each summand *)
relabel[x_] := Module[{tmp, tmp2, i},
ClearAll[cnt];(* clear the history of unique index increments *)
tmp = DeleteDuplicates@Cases[x, _V[_], Infinity];(* read all heads with indices *)
tmp2 = (newVar[#[[0]]] & /@ tmp); (* generate fresh indices for these heads *)
x /. Table[tmp[[i]] -> tmp2[[i]], {i, 1, Length[tmp]}](* substitute them in*)
]
which we use to format the output:
(* obtain lists of lists representing graphs *)
getG[k_][{a_, c__}] := Module[{fct, tmp, gr},
gr = G[k][{a, c}]; If[gr == 0, Return[0]];(* exit if no graphs generated *)
tmp = List @@ gr;(* generate list of perturbative graphs *)
fct = tmp /. Ε[__] -> 1;(* set all edges to 1 to get overall factor*)
tmp = (Flatten[(List @@ #) /. Ε[x__]^n_ :> Table[Ε[x], n]]) & /@ (tmp/ fct);(*divide out overall factors and create list of edges for each term*)
tmp = (relabel /@ tmp) /. x[V[y_][z_]] -> V[y][z];(* simplify unique vertex labels in each term *)
Transpose[{fct, tmp}](* output *)
]
Finally, if we want to show the output in terms of graphs, we can use
(* Converts the output to a list of graphs *)
ClearAll[getGraphs]
getGraphs[input_] := Module[{},
Table[
{el[[1]], Graph[((el[[2]]) /. Ε[x_, y_] -> UndirectedEdge[x, y]), VertexLabels -> "Name"]}
, {el, input}]
]
Then one can get e.g. all connected graphs as
out = getG[4][Range[4]];
SortBy[Cases[getGraphs[out], _?(ConnectedGraphQ[#[[2]]] &)], LeafCount]