I learned a long time ago that NDSolve can often be sped up by vectorizing systems of equations. I've got a problem now that seems ideal for this approach, but I can't get it to work efficiently. Any ideas on how to speed this up? This is a simplified version of my real problem, which is orders-of-magnitude larger, but has the same issues.
There are actually two related models. The first is the master equation of a birth-death-migration-extinction stochastic process. It's linear and governed by a large transition matrix TM, which is a function of the immigration rate im.
Setting up the transition matrix:
r = 1; (* birth rate *)
k = 200; (* carrying capacity *)
e = 0.01; (* extinction rate *)
m = 0.01; (* emigration rate *)
tmax = 10^6; (* maximum time *)
SetSystemOptions["SparseArrayOptions" -> "TreatRepeatedEntries" -> Total];
SetSystemOptions["NDSolveOptions" -> "DefaultSolveTimeConstraint" -> 1200];
diagonal[vec_] := Transpose[{vec + 1, vec + 1}];
superdiagonal[vec_] := Transpose[{vec + 2, vec + 1}];
subdiagonal[vec_] := Transpose[{vec, vec + 1}];
toprow[vec_] := Transpose[{ConstantArray[1, Length[vec]], vec + 1}];
from1[n_] := -r n - r n^2/k - m n - e; (* leaving n *)
dec1[n_] := r n^2/k + m n; (* going to n-1 *)
nmax = 400; (* maximum population size *)
nrange = Range[0., nmax];
transitions = Join[diagonal[nrange], subdiagonal[Rest@nrange], superdiagonal[Most@nrange], toprow[nrange]];
TM[im_] := Module[{rates},
rates = Join[
from1[nrange] - im,
dec1[Rest@nrange],
r*Most@nrange + im,
ConstantArray[e, nmax + 1]
];
SparseArray[transitions -> rates]
];
This linear master equation for the probability p of having n individuals can be quickly solved by NDSolve in vector form for a fixed immigration rate im:
(* initial probability distribution *)
p0 = Table[If[n == Round[k], 1, 0], {n, 0, nmax}];
(* 0.03 s *)
s = NDSolve`ProcessEquations[{p'[t] == TM[0.01].p[t], p[0] == p0}, p, t][[1]];
(* 0.62 s *)
NDSolve`Iterate[s, tmax];
sol = NDSolve`ProcessSolutions[s];
ListPlot[p[tmax] /. sol, PlotRange -> All]
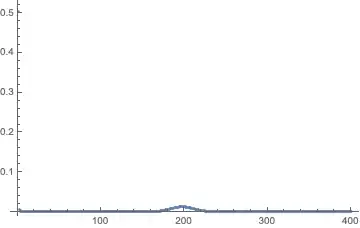
Good so far. Now the real problem: I want to make the immigration rate im a function of the probability distribution p[t] (a structured metapopulation similar to Metz & Gyllenberg 2001). This makes the system nonlinear and apparently impossible to solve efficiently in vectorized form.
First attempt:
s = NDSolve`ProcessEquations[{p'[t] == TM[m p[t].Range[0, nmax]].p[t],
p[0] == p0}, p, t][[1]];
(* NDSolve`ProcessEquations::ndnum
Encountered non-numerical value for a derivative at t == 0.`. *)
This seems wrong, since the initial immigration rate im=m p[0].Range[0, nmax] is in fact numerical:
m p0.Range[0, nmax]
(* 2. *)
Wrapping the SparseArray TM in Normal makes it work, but slowly:
(* 0.15 s *)
s = NDSolve`ProcessEquations[{p'[t] == Normal@TM[m p[t].Range[0, nmax]].p[t],
p[0] == p0}, p, t][[1]];
(* 8.26 s *)
NDSolve`Iterate[s, tmax];
sol = NDSolve`ProcessSolutions[s];
ListPlot[p[tmax] /. sol, PlotRange -> All]
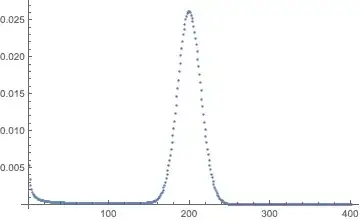
Maybe that's as good as it's going to get? No -- here are two different implementations of an unvectorized version that are faster.
First, using built-in NDSolve`ProcessEquations:
rhs = TM[m*Sum[n p[n][t], {n, 0, nmax}]].Table[p[n][t], {n, 0, nmax}];
eqns = Thread[Table[p[n]'[t], {n, 0, nmax}] == rhs];
ics = Table[p[n][0] == If[n == k, 1, 0], {n, 0, nmax}];
unks = Table[p[n], {n, 0, nmax}];
(* 2.75 s *)
s = NDSolve`ProcessEquations[Flatten[{eqns, ics}], unks, t][[1]];
(* 2.73 s *)
NDSolve`Iterate[s, tmax];
sol = NDSolve`ProcessSolutions[s];
ListPlot[Table[p[n][tmax], {n, 0, nmax}] /. sol, PlotRange -> All]
(* same output as above *)
Second, using my homemade ProcessFirstOrderODEs:
rhs = TM[m*Sum[n p[n], {n, 0, nmax}]].Table[p[n], {n, 0, nmax}];
ics = Table[If[n == k, 1, 0], {n, 0, nmax}];
unks = Table[p[n], {n, 0, nmax}];
(* 0.55 s *)
s = ProcessFirstOrderODEs[unks, rhs, ics, 0];
(* 2.82 s *)
NDSolve`Iterate[s, tmax]
sol = NDSolve`ProcessSolutions[s];
ListPlot[Table[p[n][tmax], {n, 0, nmax}] /. sol, PlotRange -> All]
(* same output as above *)
Finally, both NDSolve`Iterate stages can be sped up to around 0.6 s by processing the equations with option Jacobian -> FiniteDifference (not shown).
To recap:
- the vectorized linear problem works fine with
NDSolve - the vectorized nonlinear problem does not work with
NDSolve, except when usingNormal, which results in a total timing of 8.41 s - the nonvectorized nonlinear problem with
NDSolveis faster (5.48 s) - the nonvectorized nonlinear problem with my
ProcessFirstOrderODEsis even faster (3.37 s) - the nonvectorized nonlinear problem with my
ProcessFirstOrderODEsandJacobian->FiniteDifferenceis fastest (1.1 s)
Questions:
- can the vectorized nonlinear problem be made to work with
NDSolvewithout usingNormal? - is there any other way to speed up this
NDSolve?

?NumericQbut evidently missed the right combination. That answers my first question about an elegant, compact vectorized implementation. Too bad it's not faster than doing it as separate first-order equations. But here's anotherMethodthat brings theNDSolve`Iteratedown to 0.23s:Method -> {"IDA", "ImplicitSolver" -> {"Newton", "LinearSolveMethod" -> {"Band", "BandWidth" -> {2, 2}}}}. Decent! – Chris K Aug 04 '19 at 13:55