Suppose we have an InterpolatingFunction from NDSolve
L = 5;
sol = First@NDSolve[{D[u[x, t], t] == D[u[x, t], {x, 2}], u[x, 0] == 0,
u[-5, t] == Sin[t], u[5, t] == 0}, u, {x, -L, L}, {t, 0, 10}]
Defining the functions:
int1st[x_?NumericQ, t_?NumericQ] := NIntegrate[D[Evaluate[u[s, t] /. sol], s]*Cot[(π (x - s))/(2*L)], {s, -L, x, L}, Method -> "PrincipalValue"];
intNest[x_?NumericQ, t_?NumericQ] := NIntegrate[D[Evaluate[u[xp, t] /. sol], xp]*int1st[xp, t]*Cot[(π (x - xp))/(2*L)], {xp, -L, x, L}, Method -> "PrincipalValue"];
which are, respectively, similar to the circular Hilbert transform of a periodic function and its nest (please see the section 'Hilbert transform on the circle' on that webpage). Note in intNest, int1st is called with xp as its argument, which acts as the variable of integration of intNest.
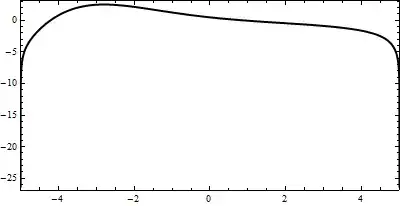
For example, plotting int1st at tn=5; I obtain the curve within 25s.
tn=5;
Plot[int1st[x,tn], {x, -L, L}, PlotRange -> {{-L, L}, All}, ImageSize -> 400, PlotPoints -> 60, AspectRatio -> 0.5, Frame -> True, Axes -> False, PlotStyle -> {Black, Thick}]
My problem is on intNest: when plotting it, there is no warning but my computer runs for hours without output. Is my definition of the intNest correct. If correct, how to make it more efficient?
Plot[intNest[x, tn], {x, -L, L}, PlotRange -> {{-L, L}, All}, ImageSize -> 400, PlotPoints -> 60, AspectRatio -> 0.5, Frame -> True, Axes -> False, PlotStyle -> {Black, Thick}]
After reading this post, I modified the function definitions:
ClearAll[int1st, intNest];
Block[{x, t}, int1st[x_?NumericQ, t_?NumericQ] = NIntegrate[D[u[s, t] /. sol, s]*Cot[(π (x - s))/(2*L)], {s, -L, x, L}, Method -> "PrincipalValue"]];
Block[{x, t}, intNest[x_?NumericQ, t_?NumericQ] = NIntegrate[D[u[xp, t] /. sol, xp]*int1st[xp, t]*Cot[(π (x - xp))/(2*L)], {xp, -L, x, L}, Method -> "PrincipalValue"]];
Findings: int1st can be plotted well in about 20s (same result as above), but it is still very slow to plot intNest.

int1st2to take advantage of the recycled points? – Chris K Feb 15 '20 at 00:50intNest[4, 5]. Rather doubt it would help withPlot, either. There aren't many integration rules that recycle points. But if the same call tointNestneeds to be done multiple times, then obviously memoization would speed things up here. – Michael E2 Feb 15 '20 at 03:21int1st2[x_?NumericQ, t_?NumericQ] := int1st2[x, t] =allowsPlot[intNest2[x, 5], ..., PlotPoints -> 5]to run in four minutes, whereas without memoizing it doesn't complete in at least fifteen minutes. – Chris K Feb 15 '20 at 14:55f[x_?NumericQ, t_?NumericQ] := f[x, t] =...can memorize all the data off[x,t]even though it is a function ofxandt? – lxy Feb 15 '20 at 15:18f[x, t]is called for a particularxandt, the value is stored for future use, called memoization (yeah, weird name). See my most popular question ever for more info. – Chris K Feb 15 '20 at 15:24intNest2[]. The key is explained in "Update 2." – Michael E2 Feb 15 '20 at 16:40intNest2, I got the warnings:>> Nonatomic expression expected at position 1 in Total[...] and obtained an empty plot. – lxy Feb 16 '20 at 10:39Total[]was an unnecessary relic, so I removed it. Perhaps you have a latent definition, which is a common reason one user gets an error and another does not. Restarting the kernel usually fixes such a problem. I doubt it's a version issue, but I'm using 12.0.0. – Michael E2 Feb 16 '20 at 12:46+dux ((2*L) Log[(L + x)/(L - x)])/\[Pi]adds the PV integral of the subtracted part. Just one last question: could u explain why, e.g. by-dux (2*L)/(\[Pi] (x - s))we subtract the singularity? – lxy Feb 16 '20 at 15:39du, $f(0)$ todux, $a$ to-2 Pi/L, and $t$ tos - x(sis the independent variable). I guess thePiecewisefunction should have{f'(0)\over a}as the default instead of zero. It's just a single point, but in a discrete algorithm, it might be an unlikely bug waiting to happen; withs == x(that is, $t=0$) specified as a singularity, it should be ok, though. – Michael E2 Feb 17 '20 at 13:47{xcoords, tcoords} = u["Coordinates"] /. sol;, are not used in the subsequent computation. What is it use? – lxy Mar 10 '20 at 06:40