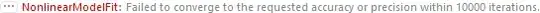I asked this question. Answers and comments were appreciated.
But (some) of the comments did not make satisfied. [Maybe due to my bad English I did not describe my problem very well].
Let me try again with another way of explanation, and I hope you can help me:
- Start with the following example:
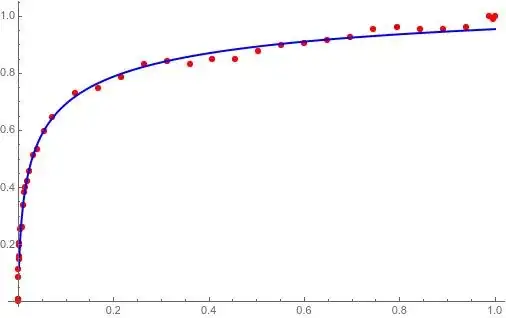
When we have these data points: $(0,5),(1,7),(2,9),(3,11)$ then Mr.A asked us to find the best fit of these points that is of the form $y=ax+b$. Then we can tell Mr.A: "the equation that you look for is $y=2x+5$".
Another person, Mr.B, asked us to find the best fit (of the same data points) that is of the form $y=ax^2+bx$. Then we should not say: "it is not good fit because the points represent a straight line, but your equation form is quadratic, and therefore we can not find because it is bad".
Yes, we can suggest Mr.B to go for straight line, but if he was strict, then we can tell him: "the equation that you look for is $y=-1.31579x^2+7.52632x$"
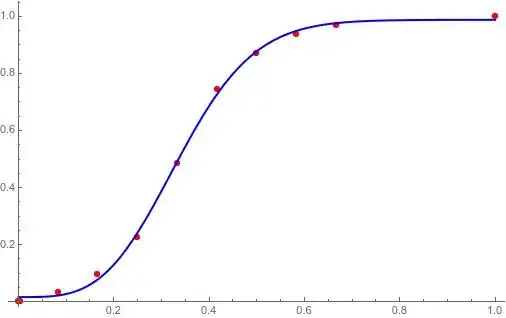
Again Mr.C asked us to find the best fit (of the same data points) that is of the form $y=ax^2+\frac{b}{x+c}$, then we can tell him: "the equation that you look for is $y=0.153282x^2+\frac{-38.869}{x+(-6.93418)}$".
What I want to say is; we should not force Mr.B and Mr.C to go for straight line.
Come to my original problem;
data = {{595098, 335}, {595158, 336}, {595218, 336}, {595338,
344}, {595458, 347}, {595638, 351}, {595818, 352}, {596082,
356}, {596322, 357}, {596922, 362}, {597521, 363}, {598481,
363}, {599322, 371}, {600523, 376}, {601723, 378}, {603523,
380}, {605323, 384}, {608924, 390}, {612523, 392}, {619724,
399}, {626926, 404}, {648527, 413}, {670129, 415}, {691731,
419}, {712906, 424}, {734504, 425}, {756104, 424}, {776690,
426}, {798291, 426}, {819890, 429}, {841490, 431}, {863090,
432}, {884692, 433}, {906290, 434}, {927892, 437}, {949492,
438}, {971090, 437}, {992691, 437}, {1014291, 438}, {1035891,
442}, {1039491, 441}, {1041236, 442}};
model = a*x + b;
fit = FindFit[data, model, {a, b}, x];
Show[Plot[Evaluate[model /. fit], {x, 595070, 1041236}],
ListPlot[data, PlotStyle -> Red]]
is fine
data = {{595098, 335}, {595158, 336}, {595218, 336}, {595338,
344}, {595458, 347}, {595638, 351}, {595818, 352}, {596082,
356}, {596322, 357}, {596922, 362}, {597521, 363}, {598481,
363}, {599322, 371}, {600523, 376}, {601723, 378}, {603523,
380}, {605323, 384}, {608924, 390}, {612523, 392}, {619724,
399}, {626926, 404}, {648527, 413}, {670129, 415}, {691731,
419}, {712906, 424}, {734504, 425}, {756104, 424}, {776690,
426}, {798291, 426}, {819890, 429}, {841490, 431}, {863090,
432}, {884692, 433}, {906290, 434}, {927892, 437}, {949492,
438}, {971090, 437}, {992691, 437}, {1014291, 438}, {1035891,
442}, {1039491, 441}, {1041236, 442}};
model = a*x^2 + b/x;
fit = FindFit[data, model, {a, b}, x];
Show[Plot[Evaluate[model /. fit], {x, 595070, 1041236}],
ListPlot[data, PlotStyle -> Red]]
is fine
data = {{595098, 335}, {595158, 336}, {595218, 336}, {595338,
344}, {595458, 347}, {595638, 351}, {595818, 352}, {596082,
356}, {596322, 357}, {596922, 362}, {597521, 363}, {598481,
363}, {599322, 371}, {600523, 376}, {601723, 378}, {603523,
380}, {605323, 384}, {608924, 390}, {612523, 392}, {619724,
399}, {626926, 404}, {648527, 413}, {670129, 415}, {691731,
419}, {712906, 424}, {734504, 425}, {756104, 424}, {776690,
426}, {798291, 426}, {819890, 429}, {841490, 431}, {863090,
432}, {884692, 433}, {906290, 434}, {927892, 437}, {949492,
438}, {971090, 437}, {992691, 437}, {1014291, 438}, {1035891,
442}, {1039491, 441}, {1041236, 442}};
model = a + b*x + c*x^2 + d*x^3 + e/x^4;
fit = FindFit[data, model, {a, b, c, d, e}, x];
Show[Plot[Evaluate[model /. fit], {x, 595070, 1041236}],
ListPlot[data, PlotStyle -> Red]]
is fine too.
But this is not fine:
data = {{595098, 335}, {595158, 336}, {595218, 336}, {595338,
344}, {595458, 347}, {595638, 351}, {595818, 352}, {596082,
356}, {596322, 357}, {596922, 362}, {597521, 363}, {598481,
363}, {599322, 371}, {600523, 376}, {601723, 378}, {603523,
380}, {605323, 384}, {608924, 390}, {612523, 392}, {619724,
399}, {626926, 404}, {648527, 413}, {670129, 415}, {691731,
419}, {712906, 424}, {734504, 425}, {756104, 424}, {776690,
426}, {798291, 426}, {819890, 429}, {841490, 431}, {863090,
432}, {884692, 433}, {906290, 434}, {927892, 437}, {949492,
438}, {971090, 437}, {992691, 437}, {1014291, 438}, {1035891,
442}, {1039491, 441}, {1041236, 442}};
model = a + b*Exp[c*(x^d + e)^f];
fit = FindFit[data, model, {a, b, c, d, e, f}, x];
Show[Plot[Evaluate[model /. fit], {x, 595070, 1041236}],
ListPlot[data, PlotStyle -> Red]]
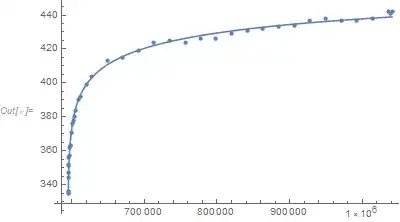
I am sure that my model is fine to represent these data points. When I re-scaled the data (dividing x by 1000000 , and dividing y by 100), DESMOS found the parameters, this means my model is representative for my original data! https://i.stack.imgur.com/6yFiZ.png
@JimB said one of my parameters is redundant, and he was right. I want to stick with
model = a + b*Exp[c*(x^d + e)^f]
I am a new user of Mathematica, please just suggest me how to rescale my data if that will work, or suggest me to use good "starting values", or suggest me to use other codes. Do not suggest me to use other models like suggesting Mr.B and Mr.C.
Edit: Desmos result of the original (not scaled data):
Your help would be really appreciated. Thanks!