ClearAll[f, xMargin, yMargin, ppX, ppY]
f[x_, y_] := Exp[-2 (x^2 + y^2)] HermiteH[2, Sqrt[2] x]^2
xMargin[x_] = Integrate[f[x, y], {y, -Infinity, Infinity}];
yMargin[y_] = Integrate[f[x, y], {x, -Infinity, Infinity}];
xrange = {-3, 3};
yrange = {-2, 2};
scale = 1/4/Pi;
gap = 0.05;
dp = DensityPlot[f[x, y], {x, xrange[[1]], xrange[[2]]}, {y, yrange[[1]], yrange[[2]]},
PlotRange -> All]
We can construct appropriately translated margins using ParametricPlot:
ppY = ParametricPlot[{xrange[[1]] - gap - scale v yMargin[y], y},
{y, yrange[[1]], yrange[[2]]}, {v, 0, 1},
PlotStyle -> Red, PlotPoints -> 50, Axes -> False];
ppX = ParametricPlot[{x, yrange[[1]] - gap - scale v xMargin[x] },
{x, xrange[[1]], xrange[[2]]}, {v, 0, 1},
PlotStyle -> Blue, PlotPoints -> 50, Axes -> False];
and combine them with dp using Show:
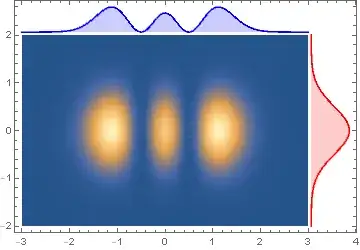
Show[ppY, ppX, dp, PlotRange -> All, Frame -> True]

To show the marginal plots on top and right frames:
ppY2 = ParametricPlot[{xrange[[2]] + gap + scale v yMargin[y], y},
{y, yrange[[1]], yrange[[2]]}, {v, 0, 1},
PlotStyle -> Red, PlotPoints -> 50, Axes -> False];
ppX2 = ParametricPlot[{x, yrange[[2]] + gap + scale v xMargin[x]},
{x, xrange[[1]], xrange[[2]]}, {v, 0, 1},
PlotStyle -> Blue, PlotPoints -> 50, Axes -> False];
Show[ppY2, ppX2, dp, PlotRange -> All, Frame -> True]

To put the marginal plots outside the frame, we can use Inset + Epilog:
insetY = Inset[#, {xrange[[2]] (1 + gap), yrange[[2]]},
{Left, Top}, Scaled[1]] & @ ppY2;
insetX = Inset[#, {xrange[[2]], yrange[[2]] (1 + gap)},
{Right, Bottom}, Scaled[1]] & @ ppX2;
Show[dp, Epilog -> {insetX, insetY},
ImagePadding -> {{Scaled[.02], Scaled[.1]}, {Scaled[.02], Scaled[.1]}},
ImageSize -> Large, PlotRangeClipping -> False, ]

Alternatively, we can Plot the functions xMargin and yMargin and use GeometricTransformation with appropriate transformation functions position them and Show the transformed graphics objects with dp:
ClearAll[transform, tFX, tFY]
transform[tf_] := Graphics[#[[1]] /.
ll : (_Line | _Polygon) :> GeometricTransformation[ll, tf]] &;
tFY = TranslationTransform[{-gap, xrange[[1]]}]@*
RotationTransform[Pi/2, {xrange[[1]], 0}];
tFX = TranslationTransform[{0, yrange[[1]] - gap}]@*
ScalingTransform[{1, -1}];
pltY = Plot[scale yMargin[y], {y, yrange[[1]], yrange[[2]]},
Filling -> Axis, PlotStyle -> Red, Axes -> False];
pltX = Plot[scale xMargin[x], {x, xrange[[1]], xrange[[2]]},
Filling -> Axis, PlotStyle -> Blue, Axes -> False];
Show[transform[tFY]@pltY, transform[tFX]@pltX, dp, PlotRange -> All,
Frame -> True]

To show the marginal plots on top and right frames use the transformations tFX2 and tFY2:
tFY2 = TranslationTransform[{gap, xrange[[1]]}]@*
RotationTransform[-Pi/2, {xrange[[2]], 0}];
tFX2 = TranslationTransform[{0, yrange[[2]] + gap}];
Show[transform[tFY2] @ pltY, transform[tFX2] @ pltX, dp, PlotRange -> All,
Frame -> True]
Update: An alternative approach to get the marginal plots: Use Plot3D to plot f with equally spaced mesh lines in x and y directions and extract the coordinates of mesh lines.
ndivs = 50;
{meshx, meshy} = Subdivide[#[[1]], #[[2]], ndivs] & /@ {xrange, yrange};
coords = Plot3D[f[x, y],
{x, xrange[[1]], xrange[[2]]}, {y, yrange[[1]], yrange[[2]]},
PlotRange -> All, Mesh -> {meshx, meshy}, PlotStyle -> None][[1, 1]];
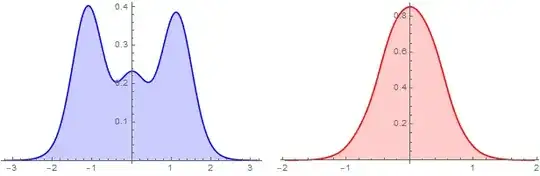
Group coords by the first and second coordinates and construct two WeightedData objects and plot them using SnoothHistogram:
bw = .01;
{wDx, wDy} = Table[Apply[WeightedData] @ Transpose @ KeyValueMap[List] @
KeySort @ GroupBy[coords, Round[#[[i]], bw] & -> Last, Mean], {i, 2}];
{sHx, sHy} = {SmoothHistogram[wDx, PlotStyle -> Blue,
Filling -> Axis, ImageSize -> 300],
SmoothHistogram[wDy, PlotStyle -> Red, Filling -> Axis, ImageSize -> 300]};
Row[{sHx, sHy}, Spacer[10]]

Alternatively, Plot the PDF of SmoothKernelDistribution of wDx and wDy:
{sKDx, sKDy} = SmoothKernelDistribution /@ {wDx, wDy};
{sHx2, sHy2} = {Plot[PDF[sKDx]@x, {x, xrange[[1]], xrange[[2]]},
PlotStyle -> Blue, Filling -> Axis, ImageSize -> 300],
Plot[PDF[sKDy]@y, {y, xrange[[1]], yrange[[2]]}, PlotStyle -> Red,
Filling -> Axis, ImageSize -> 300]};
Row[{sHx2, sHy2}, Spacer[10]]

Update 2: Processing DensityPlot output to get {x,y,z} coordinates (where z is scaled to the unit interval:
dp = DensityPlot[f[x, y], {x, -3, 3}, {y, -2, 2},
ColorFunction -> Hue, PlotRange -> All, PlotPoints -> 50]
coordsFromDP = Join[dp[[1, 1]], List /@ dp[[1, 3, 2, All, 1]], 2];
Except for the scale of the z coordinate ListPlot3D of coordsFromDP is "close" to the Plot3D output:
Row @ {Plot3D[f[x, y], {x, -3, 3}, {y, -2, 2}, ImageSize -> 300,
PlotRange -> All], ListPlot3D[coordsFromDP, ImageSize -> 300]}

We process coordsFromDP the same way we did for coords above (except for a larger bin width):
bw = .02;
{wDx2, wDy2} = Table[Apply[WeightedData] @ Transpose @ KeyValueMap[List] @
KeySort@GroupBy[coordsFromDP, Round[#[[i]], bw] & -> Last, Mean], {i, 2}];
{sHx2, sHy2} = {SmoothHistogram[wDx2, PlotStyle -> Blue,
Filling -> Axis, ImageSize -> 300],
SmoothHistogram[wDy2, PlotStyle -> Red, Filling -> Axis, ImageSize -> 300]};
Row[{sHx2, sHy2}, Spacer[10]]










f[x,y]is expensive to compute, and you don't want to have to keep recalling it for the density plot and the 1D plots, you have two choices. First, you could use memoization viaf[x_, y_] := f[x,y] = <expensive computation>. Alternatively you could precompute the density as a matrix of values and callListDensityPloton it. – Jason B. Sep 10 '20 at 16:24ListDensityPlotis ~3x slower for similar resolution, which I chalked up to the adaptive mesh ofDensityPlot, which I'd like to keep. Do you know how to extract the data (i.e., the (x,y,z) coordinates) underlyingDensityPlot? If so, memoization would work really well in combination with @kglr 's answer below. – Jaffe42 Sep 10 '20 at 17:41