The only thing that can be parallelized here is the generation of the interpolation points. For example:
Clear[integrate, interpolate];
integrate[f_, r2_?NumericQ, a_?NumericQ] := NIntegrate[
r1 f[r1] BesselJ[0, r1 r2] E^(-I (r1^2 + r2^2)/2),
{r1, 0, a}
];
interpolate[f_, a_?NumericQ, step_?NumericQ] := Interpolation[
ParallelTable[
{r2, integrate[f, r2, a]},
{r2, 0, a, step}
]
];
Iterate the procedure twice, starting with a constant function:
results = NestList[
interpolate[#, 1, 0.01] &,
1 &,
2
];
Plot the obtained functions at each iteration:
ReImPlot[Evaluate[Through[results[r2]]],
{r2, 0, 1},
PlotLegends -> Range[0, Length[results] - 1]
]
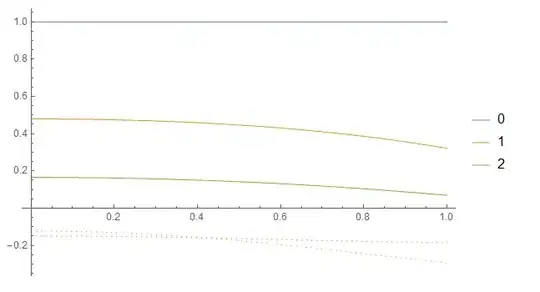
Edit
I don't think that with NIntegrate you can speed this up any further. The free parameter r2 appears in the body of the integral, so I don't think that interpolation is going to help you too much here. You could make an interpolation over both r1 and r2 of which you then take the antiderivative over r1, but you also risk introducing numerical error that way. There's probably a reason why NIntegrate isn't faster here.
I do have another idea for speeding this up, though. We can use ParametricNDSolveValue to do the integration instead by re-phrasing the problem and considering r2 to be a parameter. This should reduce the top-level overhead involved in calling NIntegrate multiple times. Here's how to do it:
Clear[integrate, interpolate];
integrate[f_, a_?NumericQ] := ParametricNDSolveValue[
{y'[r1] == r1 f[r1] BesselJ[0, r1 r2] E^(-I (r1^2 + r2^2)/2), y[0] == 0},
y[a],
{r1, 0, a},
{{r2, 0, a}}
];
interpolate[f_, a_?NumericQ, step_?NumericQ] := With[{
int = integrate[f, a]
},
Interpolation[
ParallelTable[
{r2, int[r2]},
{r2, 0, a, step},
Method -> "CoarsestGrained"
]
]
];
results = NestList[interpolate[#, 1, 0.01]&, 1&, 20];
ReImPlot[
Evaluate[Through[results[r2]]],
{r2, 0, 1},
PlotLegends -> Range[0, Length[results] - 1], PlotRange -> All
]
This should be significantly faster than the method based on NIntegrate. You can also take a look at FunctionInterpolation to do the Interpolation step, since this might save you from having to sub-sample the integral more than you really need. For example:
int = Quiet @ FunctionInterpolation[
integrate[1&, 1][r2],
{r2, 0, 1},
InterpolationPoints -> 50
]
ReImPlot[int[r2], {r2, 0, 1}]
FunctionInterpolation cannot be parallelized, though, so it's a trade-off

r2but not the integration step itself. Any advice on this? – user135626 Nov 17 '20 at 23:36r2variable)? Or is your coding method faster? – user135626 Nov 18 '20 at 00:13ParallelTableis better here thanParallelDo. – Sjoerd Smit Nov 18 '20 at 14:13Withcommand in your answer to solve the paramteric ODE before ther2parameter values are assigned inParallelTableas that would cause error inParametricNDSolveValue? (3) But isn'tParallelDoalso assigning differentr2values to different kernels, just asParallelDowould do here? – user135626 Nov 19 '20 at 04:15WithensuresParametricNDSolveValueevaluates only once, thus cutting down on redundant operations. – Sjoerd Smit Nov 19 '20 at 09:09f2[r2]do not exist in the master kernel. You can useSetSharedVariables[f2], before theParallelDo, but then each assignment will have to be transmitted to all other kernels (master and slave), causing communication overhead. In general, I recommend always usingParallelTableandParallelEvaluatewhenever you can. There aren't many situations where you needParallelDoand you need a good understanding of the mechanisms involved to use it. – Sjoerd Smit Nov 19 '20 at 09:15