all the molecules have the same orientation at any time, and I think I see why
Because at each timestep you are are computing only a single angle and using it to rotate every molecule.
In this approach I use NestList to accumulate a list of TransformationFunction objects. The key here is that Composition[TransformationFunction[..], TransformationFunction[...]] will evaluate to a single transformation. So at each step I take the previous transformation and compose it with a random translation and a random rotation.
randomInitialTransform[] := Composition[
TranslationTransform[RandomReal[{-5, 5}, 3]],
RotationTransform[RandomReal @ {-Pi, Pi}, RandomReal[{-1,1}, 3]]
];
randomStep[inputTransform_] := Composition[
(*small random translation*)
TranslationTransform @ RandomReal[{-0.1, 0.1}, 3],
(*small random rotation*)
RotationTransform[RandomReal @ {-0.1, 0.1}, RandomReal[{-1,1}, 3]],
inputTransform
]
randomTrajectory[n_] := NestList[randomStep, randomInitialTransform[], n];
Now make a table of trajectories, and grab the GraphicsComplex from a molecule plot:
trajectoryList = Table[randomTrajectory @ 200, 30];
graphicsComplex = First @ MoleculePlot3D[Molecule @ "water", PlotTheme -> "Tubes"];
The next part is to use GeometricTransformation, and give a list of transforms as the second argument. For example to visualize all steps of a single trajectory use
Graphics3D[GeometricTransformation[graphicsComplex, randomTrajectory[200]]]
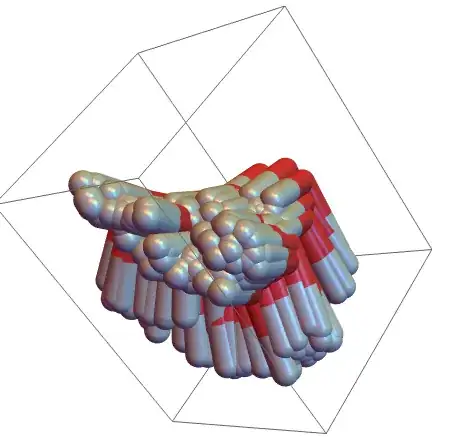
To visualize a snapshot of the ensemble use something like
snapShot[n_] := Graphics3D[
GeometricTransformation[graphicsComplex, trajectoryList[[All, n]]],
PlotRange -> {{-6, 6}, {-6, 6}, {-6, 6}}
]


