I'm trying to develop a continuous time unscented Kalman filter for a dynamic system inspired to this article: "On Unscented Kalman Filtering for State Estimation of Continuous-Time Nonlinear Systems" of the author Simo Särkkä. In particular the Square Root one.
My systems has dimension 5 so I have a system of 11 x 5 + 15 non linear coupled first order differential equations.
The code of my notebook is very long and laborious, in the end I simulate a dynamic system and the filter receive the output.
When I try to solve the system with NDSolve it says:

So I do as recommended and it works till it find a singularity that I think is just numerical and not due to the system.
Maybe using Chop to simplify the approximations as I saw in other posts.
This is a short versione of my code:
t0 = 0;
tfin = 4 Pi;
pstl = {xstl, ystl};
R0l[\[Theta]_] = {{Cos[\[Theta]], -Sin[\[Theta]]} , {Sin[\[Theta]],
Cos[\[Theta]]}};
pmk1 = {xmk1, ymk1};
pmk2 = {xmk2, ymk2};
numericalvalues =
Thread[{l1, l2, xstl, ystl, xmk1, ymk1, xmk2, ymk2} -> {8, 5,
3, -2, 32, 16, -15, 2}];
qalist = {x, y, [Theta], xstls, ystls};
qa[t_] = (ToString[#] <> "[t]") & /@ qalist // ToExpression;
qad[t_] = D[qa[t], t];
qadd[t_] = D[qa[t], t];
v[t_] = 2;
[Omega][t_] = 1;
input[t_] = {v[t], [Omega][t]};
g1 = {Cos[#], Sin[#], 0, 0, 0} & @(#[[3]]) &;
g2 = {0, 0, 1, 0, 0};
dyn = (g1@#v[t] + g2[Omega][t]) &;
pstf = ({#1, #2} + R0l[#3 ].{#4, #5}) &;
h1 = (Sqrt[#.#] &[pmk1 - pstf @@ #]) &;
h2 = (Sqrt[#.#] &[pmk2 - pstf @@ # ]) &;
CreateWhiteNoise[[Mu], s, t0_, tfin_, dt_, p_] :=
Module[{distr, n = Length[[Mu]], listt, nc, listrn},
listt = Range[t0, tfin, dt];
nc = Length[listt];
distr = MultinormalDistribution[[Mu], s];
listrn = RandomVariate[distr, nc];
(Interpolation[Thread[Join[{listt, listrn[[All, #]]}]]][p]) & /@
Range@n
] (Just create white noise continuous time sample)
cov = {{0.008, 0}, {0,
0.008}}; ((0.008//Sqrt) 3 [Equal] 0.268 [m]*)
media = {0, 0};
noise = CreateWhiteNoise[media, cov, t0, tfin + 0.1, 0.04, t ];
h = {h1@#, h2@#} & ;
hn = (h@# + noise) &;
output[t_] = Join[h@qa[t], hn@qa[t]] /. numericalvalues;
eqdyn = qad[t] == dyn@qa[t];
qa0 = Flatten@{-2, 1, Pi/3, pstl} /. numericalvalues;
eqin = qa[0] == qa0;
monitor = {qa[t], input[t], output[t]};
{state, in, out} =
NDSolveValue[{eqdyn, eqin}, monitor, {t, t0, tfin}];
ParametricPlot[state[[1 ;; 2]], {t, t0, tfin}]
And I get the desired result:
If now I want to use the filter as in the paper:
qlisthat = (ToString@# <> "hat" // ToExpression) & /@ qalist ;
qahat[t_] = (ToString@# <> "[t]" // ToExpression) & /@ qlisthat;
qahatd[t_] = D[qahat[t], t];
nstate = Length@qlisthat;
Covariance Matrix
Pmat = Array[ToExpression[("P" <> ToString[#1] <> ToString[#2])] &,
IdentityMatrix[nstate] // Dimensions];
P[t_] = Array[ToExpression[(ToString@Pmat[[#1, #2]] <> "[t]")] &,
Dimensions[Pmat]];
Pd[t_] = D[P[t], t];
Initial estimate
P0 = IdentityMatrix[nstate]*{0.005, 0.005, 0.3 Degree, 0.005,
0.005 };
stima0 = RandomVariate[MultinormalDistribution[ qa0, P0]];
initstima = qahat[0] == stima0;
initcov = P[0] == P0;
initEKF = {initstima, initcov};
FILTER PARAMETERS AND VARIABLES
k = 1;
[Alpha] = 1;
[Beta] = 0;
nsp = 2 nstate + 1;
[Lambda] = [Alpha]^2 (nstate + k) - nstate;
c = Sqrt[[Alpha]^2 (nstate + k)];
Covariance matrix factorization
decP = SparseArray[{i_, j_} /; i >= j :>
ToExpression["a" <> ToString[(10 #1 + #2) &[i, j]]] ,
Dimensions[Pmat]] // Normal;
Dec[t_] =
Array[If[decP[[#1, #2]] == 0, 0 ,
0, (ToString@decP[[#1, #2]] <> "[t]" ) // ToExpression] &,
Dimensions@Pmat];
Decd[t_] = D[Dec[t], t];
Sigma Points
XSP = SparseArray[{{i_,
j_} :> ("xsp" <> ToString@i <> ToString@j )} , {nsp,
nstate}] // Normal // Transpose;
X[t_] = Array[(ToString@XSP[[#1, #2]] <> "[t]" // ToExpression) &,
Dimensions@XSP];
Xd[t_] = D[X[t], t];
Sigma points propagation
propX = dyn /@ (X[t][Transpose]) // Transpose // Simplify;
Sigma points output
Ysp = {h1 /@ (X[t][Transpose]), h2 /@ (X[t][Transpose])} /.
numericalvalues // Simplify;
z = Transpose@D[hn /@ (X[t][Transpose]), t] /. numericalvalues;
Weights
w0 = [Lambda] / (nstate + [Lambda]);
w = 1 / ( 2 (nstate + [Lambda]));
w0' = w0 + 1 - [Alpha]^2 + [Beta];
wm = Append[{w0}, Table[w, nsp - 1]] // Flatten;
Wm = Table[wm, nsp] // Transpose;
wc = ReplacePart[wm, 1 -> w0'];
Wc = SparseArray[{{1, 1} -> w0', {i_, i_} /; i > 1 -> w},
nsp {1, 1}] // Normal;
W = (IdentityMatrix[nsp] - Wm).Wc.Transpose[
IdentityMatrix[nsp] - Wm] // Simplify;
FILTER EQUATIONS
[CapitalPhi][mat_] :=
Module[{i, j},
SparseArray[{{i_, i_} :> mat[[i, i]]/2, {i_, j_} /; i > j :>
mat[[i, j]] }, Dimensions[mat]] // Normal];
StackCols[l__?MatrixQ] := MapThread[Join, {l}];
ZeroMatrix[m_, n_] := Normal@SparseArray[{}, {m, n}];
ListForm[mat_] :=
DeleteCases[(Thread@# & /@ Thread@mat // Flatten ), True]
K = X[t].W. Ysp[Transpose].Inverse[cov];
U = propX.W.X[t][Transpose] + X[t].W.propX[Transpose] -
K.cov.K[Transpose];
M = Inverse@Dec[t].U.Transpose[Inverse@Dec[t]];
stimaSRUKBF = X[t].wm;
eqDecP = Decd[t] == Dec[t].[CapitalPhi][M];
eqX = Xd[t] ==
propX.Wm + K.(z - Ysp.Wm) +
c * StackCols[ZeroMatrix[nstate, 1],
Dec[t].[CapitalPhi][M], -Dec[t].[CapitalPhi][M]];
eqSRUKBF = {ListForm@eqDecP, eqX};
INITIAL CONDITIONS
A0 = CholeskyDecomposition@P0;
X0 = Transpose[Table[stima0, nsp]] +
c * StackCols[ZeroMatrix[nstate, 1], A0, -A0];
initDecp = Dec[0] == A0;
initX = X[0] == X0;
initSRUKBF = {ListForm@initDecp, initX};
INTEGRATION
errstima = qa[t] - stimaSRUKBF;
monitorSRUKBF = {qa[t], stimaSRUKBF, errstima, Xd[t][Transpose]};
(Xd[t] are the derivatives cause singularity)
(With[{opts=SystemOptions[]},Internal`WithLocalSettings[
SetSystemOptions["NDSolveOptions"[Rule]"DefaultSolveTimeConstraint"
[Rule]10.],{stat, statoSRUKBF, errstimaSRUKBF} = NDSolveValue[{eqns,
eqin, eqSRUKBF, initSRUKBF}, monitorSRUKBF,
{t,t0,0.04}];,SetSystemOptions[opts]]])
({stat, statoSRUKBF, errstimaSRUKBF} = NDSolveValue[{eqdyn, eqin,
eqSRUKBF, initSRUKBF}, monitorSRUKBF,
{t,t0,0.04}];)
{stat, statoSRUKBF, errstimaSRUKBF, der} =
NDSolveValue[{eqdyn, eqin, eqSRUKBF, initSRUKBF}, monitorSRUKBF,
{t, t0, 0.5}, Method -> {"EquationSimplification" -> "Residual"} ];
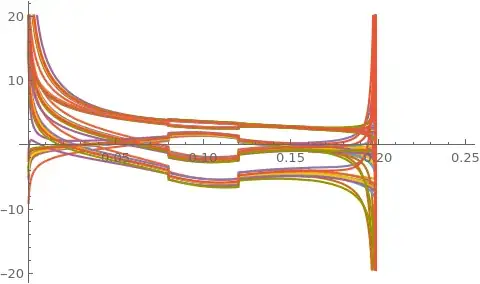
(NDSolveValue::ndsz: At t == 0.1982530364128179`, step size is effectively zero; singularity or stiff system suspected.)
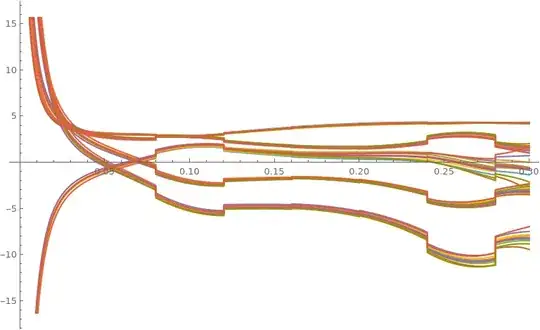
Plot[der, {t, t0, 0.25}]
The plot shows the derivatives of the variables that cause singularity. I don't know if is there a way to make the equations in a simpler form that NDSolve can handle, or if is there some trick I can use to manage singularities. The fact is that I think singularity is a numerical matter and not due to the formal system. \
EDIT Replace just these lines of code to make things work:
z = D[hn@qa[t], t] /. numericalvalues // Simplify;
Z = Table[z, nsp] // Transpose;
eqX = Xd[t] ==
propX.Wm + K.(Z - Ysp.Wm) +
c * StackCols[ZeroMatrix[nstate, 1],
Dec[t].[CapitalPhi][M], -Dec[t].[CapitalPhi][M]]
And now the system doesn't go on singularity. Before happened because the Dec[t] lost rank during system evolution, but it shouldn't happen, so I've found out the filter's equations weren't right obtaining the following results instead of the previous ones
NDSolveis called. Maybe that's the problem – Michael E2 Mar 27 '21 at 19:57NDSolvecan handle arbitrary delays (e.g.Derivative[1][R0l][xsp103[t]]), but maybe the prior errors resulted in an incorrect system. – Michael E2 Mar 27 '21 at 20:00initSRUKBFinvolves a number of0==0, please double check it. – xzczd Mar 28 '21 at 01:41ninw = 1 / ( 2 (n + \[Lambda]));is not defined. – xzczd Mar 28 '21 at 08:43