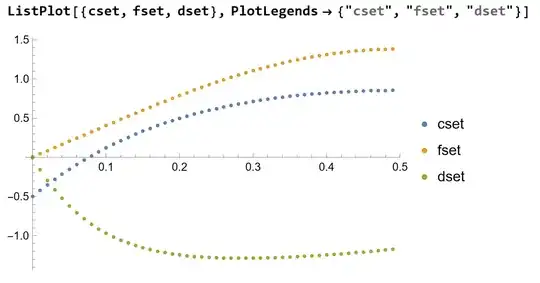
I am fitting dataset dset to a set of non-linear differential equations to find the fitting parameters dP (pressure differential) and LK (thermodynamic tension)
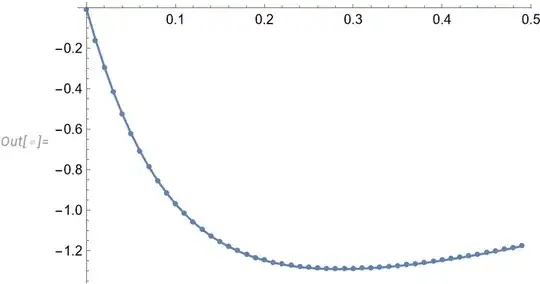
dset = {{0.0005`, -0.010556322978021847`}, {0.010500000000000006`, \
-0.16182647440121997`}, {0.020500000000000015`, \
-0.29695349218330797`}, {0.030500000000000024`, \
-0.4182634329865751`}, {0.04050000000000003`, -0.5269223967724705`}, \
{0.05050000000000004`, -0.6240338483581807`}, {0.06050000000000005`, \
-0.7106407676192987`}, {0.07050000000000005`, -0.7877204571404507`}, \
{0.08050000000000006`, -0.856181218817242`}, {0.09050000000000007`, \
-0.9168613181414874`}, {0.10050000000000008`, -0.9705298179210835`}, \
{0.11050000000000008`, -1.0178887965803456`}, {0.1205000000000001`, \
-1.059576525235709`}, {0.1305000000000001`, -1.096171257192166`}, \
{0.1405000000000001`, -1.1281953593331848`}, {0.1505000000000001`, \
-1.1561195807387954`}, {0.16050000000000011`, -1.1803673085219024`}, \
{0.17050000000000012`, -1.2013187048750018`}, {0.18050000000000013`, \
-1.219314653893368`}, {0.19050000000000014`, -1.2346604733142499`}, \
{0.20050000000000015`, -1.247629366294407`}, {0.21050000000000016`, \
-1.2584656030266932`}, {0.22050000000000017`, -1.2673874324894605`}, \
{0.23050000000000018`, -1.2745897318716903`}, {0.24050000000000019`, \
-1.2802464059943979`}, {0.25050000000000017`, -1.284512551975971`}, \
{0.2605000000000002`, -1.2875264059562497`}, {0.2705000000000002`, \
-1.2894110892831467`}, {0.2805000000000002`, -1.2902761714693076`}, \
{0.2905000000000002`, -1.2902190666663456`}, {0.3005000000000002`, \
-1.289326279545788`}, {0.3105000000000002`, -1.2876745154405251`}, \
{0.32050000000000023`, -1.2853316684758496`}, {0.33050000000000024`, \
-1.2823577002670472`}, {0.34050000000000025`, -1.2788054206235198`}, \
{0.35050000000000026`, -1.2747211806057743`}, {0.36050000000000026`, \
-1.2701454872488802`}, {0.3705000000000003`, -1.2651135483041822`}, \
{0.3805000000000003`, -1.259655754464611`}, {0.3905000000000003`, \
-1.2537981057286822`}, {0.4005000000000003`, -1.2475625878223087`}, \
{0.4105000000000003`, -1.2409675039324466`}, {0.4205000000000003`, \
-1.2340277664078854`}, {0.4305000000000003`, -1.2267551525451035`}, \
{0.44050000000000034`, -1.2191585280958825`}, {0.45050000000000034`, \
-1.2112440417030041`}, {0.46050000000000035`, -1.2030152930855527`}, \
{0.47050000000000036`, -1.1944734774498176`}, {0.48050000000000037`, \
-1.1856175082766092`}, {0.4905000000000004`, -1.1764441200214577`}};
fset = {{0.0005, 0.0029994884862318545}, {0.010500000000000006, 0.04290422551646877}, {0.020500000000000015, 0.08271521578894832}, {0.030500000000000024, 0.1225075890771862}, {0.04050000000000003, 0.16232844620349135}, {0.05050000000000004, 0.2022016182928511}, {0.06050000000000005, 0.24213173178259795}, {0.07050000000000005, 0.2821076803700469}, {0.08050000000000006, 0.32210558852161697}, {0.09050000000000007, 0.36209133779687963}, {0.10050000000000008, 0.4020227165941489}, {0.11050000000000008, 0.44185124530069814}, {0.1205000000000001, 0.4815237217048463}, {0.1305000000000001, 0.5209835255288894}, {0.1405000000000001, 0.5601717158155678}, {0.1505000000000001, 0.599027950470219}, {0.16050000000000011, 0.6374912534028201}, {0.17050000000000012, 0.6755006513403476}, {0.18050000000000013, 0.7129956994240441}, {0.19050000000000014, 0.7499169121163021}, {0.20050000000000015, 0.7862061136751125}, {0.21050000000000016, 0.8218067204737102}, {0.22050000000000017, 0.8566639657168472}, {0.23050000000000018, 0.8907250756040312}, {0.24050000000000019, 0.9239394046879505}, {0.25050000000000017, 0.9562585370494301}, {0.2605000000000002, 0.9876363589372191}, {0.2705000000000002, 1.0180291076824612}, {0.2805000000000002, 1.0473954009765578}, {0.2905000000000002, 1.075696249981966}, {0.3005000000000002, 1.1028950592146718}, {0.3105000000000002, 1.1289576156826286}, {0.32050000000000023, 1.1538520693758882}, {0.33050000000000024, 1.1775489068722165}, {0.34050000000000025, 1.200020919538734}, {0.35050000000000026, 1.2212431675685949}, {0.36050000000000026, 1.241192940885903}, {0.3705000000000003, 1.2598497177767924}, {0.3805000000000003, 1.277195121955397}, {0.3905000000000003, 1.2932128786464807}, {0.4005000000000003, 1.3078887701584572}, {0.4105000000000003, 1.3212105913286585}, {0.4205000000000003, 1.3331681051444677}, {0.4305000000000003, 1.343752998777381}, {0.44050000000000034, 1.3529588402103094}, {0.45050000000000034, 1.3607810355900682}, {0.46050000000000035, 1.3672167873955854}, {0.47050000000000036, 1.372265053476814}, {0.48050000000000037, 1.375926506987712}, {0.4905000000000004, 1.3782034971952344}};
cset = {{0.0005, -0.5042310407443639}, {0.010500000000000006, \ -0.427674601050752}, {0.020500000000000015, -0.3544091437700765},
{0.030500000000000024, -0.28474223380967734},
{0.04050000000000003, -0.21845219304523977}, {0.05050000000000004, \ -0.1553452433932613}, {0.06050000000000005, -0.09525380897089007},
{0.07050000000000005, -0.038030681942581725}, {0.08050000000000006, 0.016455923730000113}, {0.09050000000000007, 0.0683261356767569}, {0.10050000000000008, 0.11769117737156293}, {0.11050000000000008, 0.164655332408679}, {0.1205000000000001, 0.2093173133085545}, {0.1305000000000001, 0.25177119317041247}, {0.1405000000000001, 0.29210702401667615}, {0.1505000000000001, 0.33041123620685403}, {0.16050000000000011, 0.36676688931358775}, {0.17050000000000012, 0.4012538259126424}, {0.18050000000000013, 0.43394876511503755}, {0.19050000000000014, 0.4649253615923762}, {0.20050000000000015, 0.49425424760155606}, {0.21050000000000016, 0.5220030694835737}, {0.22050000000000017, 0.5482365257784656}, {0.23050000000000018, 0.5730164110488714}, {0.24050000000000019, 0.5964016674098069}, {0.25050000000000017, 0.6184484443671279}, {0.2605000000000002, 0.6392101666770944}, {0.2705000000000002, 0.6587376094076599}, {0.2805000000000002, 0.677078979099448}, {0.2905000000000002, 0.6942799998106967}, {0.3005000000000002, 0.7103840028276676}, {0.3105000000000002, 0.7254320188886919}, {0.32050000000000023, 0.7394628718770215}, {0.33050000000000024, 0.7525132730649463}, {0.34050000000000025, 0.764617915125826}, {0.35050000000000026, 0.7758095652630415}, {0.36050000000000026, 0.7861191569301358}, {0.3705000000000003, 0.7955758797315625}, {0.3805000000000003, 0.8042072671971728}, {0.3905000000000003, 0.8120392822155327}, {0.4005000000000003, 0.8190963999917696}, {0.4105000000000003, 0.8254016884657382}, {0.4205000000000003, 0.8309768861869055}, {0.4305000000000003, 0.8358424776946949}, {0.44050000000000034, 0.8400177664982754}, {0.45050000000000034, 0.8435209457891544}, {0.46050000000000035, 0.8463691670545794}, {0.47050000000000036, 0.8485786067907134}, {0.48050000000000037, 0.8501645315425272}, {0.4905000000000004, 0.8511413615178258}};
below are the equations and the model for fitting the data
c0 = -4.0;
eqn1 = c'[s] == (-2 d[s] Sqrt[1 - f[s] c[s]^2])/f[s];
eqn2 = f'[s] == 4 Sqrt[1 - f[s] c[s]^2];
eqn3 = d'[s] == -(2 c[s]^2 (d[s] - c0) + c[s] (c0^2 - d[s]^2) + LK c[s] +
dP + 4 d[s] (1 - f[s] c[s]^2)/f[s])/Sqrt[1 - f[s] c[s]^2];
system = {eqn1, eqn2, eqn3, f[0.0005] == 0.0029994884862318545, d[0.0005] == -0.010556322978021847, c[0.0005] == -0.5042310407443639`};
model = ParametricNDSolveValue[system, d, {s, 0.0005, 0.5}, {dP, LK}, MaxSteps -> Infinity];
I get an error in execution when I try to use the entire dset for fitting. And although I get the correct fitting parameter values for dP and LK, this happens when I use a subset i.e. dset[[;;ind]] where ind is less than 38; in this case when the evaluation occurs, I get a complaint that the step size effectively goes to 0 at a particular value of s.
fit = NonlinearModelFit[dset[[1 ;; 37]], model[dP, LK][s], {dP, LK}, s, Method -> "Gradient"];
(*ParametricNDSolveValue::ndsz: At s == 0.37654293376950654`, step size is effectively zero; singularity or stiff system suspected.*)
fit["BestFitParameters"]
{dP -> 17.48, LK -> -39.9968} (*these values are correct as dset, fset and cset were generated using these fitting parameters*)
My question is how to avoid the step size from reaching zero and possibly use the entire dataset dset for fitting?


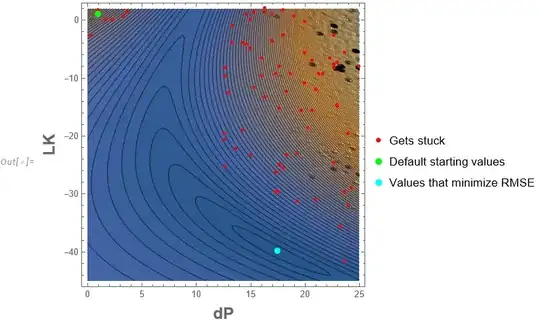
fit =NonlinearModelFit[dset.....? – bmf Feb 18 '23 at 05:28dsetNonlinearModelFit does not execute. Trydset[[;;36]– Ali Hashmi Feb 18 '23 at 07:54dsetonly, whilemodeldepends oncandfas well? – Alex Trounev Feb 18 '23 at 08:08csetandfsetbut could not get the code to work. Do you have an idea how to incorporate them as well and make the function work. Thanks ! – Ali Hashmi Feb 18 '23 at 08:11cset,fsetanddsettogether but it seems that the solver get stuck. https://mathematica.stackexchange.com/questions/28461/how-to-fit-3-data-sets-to-a-model-of-4-differential-equations – Ali Hashmi Feb 18 '23 at 09:11