ChemDraw can interpret the correct representation, even if this is not explicitly represented in the CDX file. So the idea is to remotely control ChemDraw to get the information, and to do that we have to build three pieces of functionality:
Drive ChemDraw with AppleScript to open a CDX file, select all of the molecules (there should only be one...), and Edit>Copy As...>SMILES
Retrieve the results from the clipboard and use this to populate the entry in our record (with a sanity check against the stated molecular weight and molar mass)
Automate downloading the CDX files from the URLs, putting the results into a temporary directory to process them
This type of system-level application scripting is going to be very operating system dependent, so the solution below will only work on a Mac; for the record I performed this with MacOS 13.5 and ChemDraw 22.2 (and Mathematica 13.3).
We start by creating a lightweight function wrapper around the osascript command line program which will let us run AppleScripts; this codes is copied directly from a Stack Exchange post on using AppleScript in Mathematica:
(* Code source: https://mathematica.stackexchange.com/questions/36764/how-to-import-a-numbers-spreadsheet/36772#36772 *)
AppleScript["RunFile", file_] := Run["osascript " <> file]
AppleScript["RunScript", script_] := With[
{file = ToFileName[$TemporaryDirectory, "script.txt"]},
Export[file, script, "String"];
AppleScript["RunFile", file]
]
After a bit of fumbling around, I devised the following AppleScript which takes a file path (provided as the typical POSIX filepath string) as input and results in the SMILES string on the clipboard. Once you know how to specify keystroke commands in AppleScript, it is relatively straightforward. After getting the script to run in the ScriptEditor, I created the following lightweight function wrapper using a StringTemplate:
scriptTemplate[file_] := AbsoluteFileName[file] // StringTemplate["set p to \"``\"set f to POSIX file ptell application \"ChemDraw\" open f activate tell application \"System Events\" keystroke \"a\" using {command down} keystroke \"c\" using {option down, command down} keystroke \"w\" using {command down} end tell delay 0.5end tell"]
(I found that it worked more reliably if I inserted a short delay after the copy and paste operation.) We also need to retrieve the result from the clipboard, as described in this stackoverflow thread (including versions for Windows and Ubuntu; the version implemented below is for MacOS):
(* Code source: https://mathematica.stackexchange.com/a/130224/63709 *)
Clear[getSMILESFromCDX]
getClipboard[___] := Import["!pbpaste", "Text"]
getSMILESFromCDX[file_] := getClipboard @ AppleScript["RunScript", scriptTemplate[file]]
(demo)
getSMILESFromCDX["~/Downloads/BuBTBP.cdx"]
("CCCCC(N=N1)=C(CCCC)N=C1C2=NC(C3=NC(C4=NN=C(CCCC)C(CCCC)=N4)=CC=C3)=CC=C2")
Finally, we want to be able to retrieve a file from an URL, analogous to Import. We will write a custom function using URLDownload returns a File to a temporary download directory):
importMolFromURL[url_] := Molecule @ getSMILESFromCDX @ URLDownload[url]
(demo)
url = "https://www.oecd-nea.org/ideal/structures/BuBTBP.cdx?fileKey=238";
importMolFromURL[url]


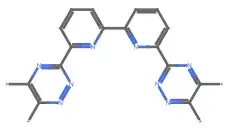

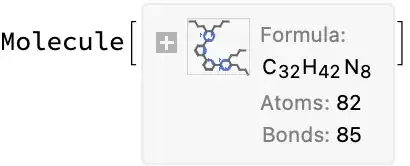
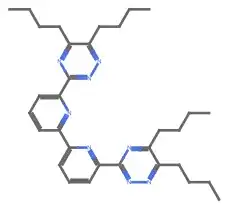

File>Export...) and then usingImportto read the resulting MOL file does seem to work correctly.(I'm also hacking out a workaround where I use AppleScript to have ChemDraw Copy-As-SMILES and then read that from the clipboard.)
– Joshua Schrier Oct 13 '23 at 22:42C4H9in the source files withnButhenMoleculewill import it correctly. – Joshua Schrier Oct 24 '23 at 02:10