I'm looking into fitting some data with Mathematica. I've got my head around how NonlinearModelFit works (I've been using the Levenberg-Marquardt algorithm for some other work).
But my data this time is Poisson distributed, and I want to see if using the appropriate MLE for Poisson data is better for my scenario than nonlinear least squares fitting.
According to the paper Efficient Levenberg-Marquardt minimization of the maximum likelihood estimator for Poisson deviates, then the minimization for least-squares fitting, for data $y_{i}$ and the model $f_{i}$ is
$$ \chi^{2}=2\sum_{i=1}^{N} \frac{\left ( f_{i}-y_{i} \right )^{2}}{\sigma_{i}^{2}} $$
whereas for Poisson distributed data according to the paper, the minimization is
$$ \chi^{2}=2\sum_{i=1}^{N}f_{i}-y_{i}-2\sum_{i=1,y\neq 0}^{N}y_{i}\ln \left ( \frac{f_{i}}{y_i} \right ) $$
Is it possible to run a model-fitting in Mathematica using this minimization? And can a (modified?) Levenberg-Marquardt algorithm still be used?
Edit
There's an associated Nature Methods letter at http://dx.doi.org/10.1038/nmeth0510-338, with a revised version of the above link: http://www.nature.com/nmeth/journal/v7/n5/extref/nmeth0510-338-S1.pdf (courtesy of @belisarius)
Update #1
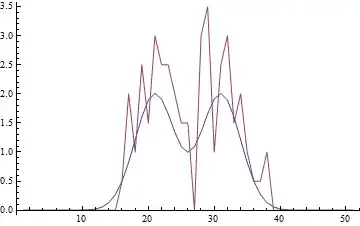
So here's the sort of data/model I'm looking to fit: the sum of two (or more) Gaussians, which may sometimes overlap as shown in the example below.
The amount of Poisson noise is deliberately significant as I'm dealing with very low counts. I've only posted a one-dimensional example here, but the data is in 2D, so there are more variables (x,y,means,heights,sigma...). I'm happy with using NonlinearModelFit to solve the problem, but I'm curious about dealing with the Poisson noise "more appropriately".
twoGaussianFunction[x_, A1_, sigma1_, mean1_, A2_, sigma2_, mean2_] :=
A1 Exp[-((x - mean1)^2/(2 sigma1^2))] +
A2 Exp[-((x - mean2)^2/(2 sigma2^2))];
cleandata = Table[twoGaussianFunction[i, 10, 10, 30, 10, 10, 60], {i, 0, 100}];
noisydata = RandomVariate[PoissonDistribution[0.5 #]] & /@ cleandata;
ListLinePlot[{cleandata, noisydata}, PlotRange -> Full]


findfitfunction setting theNormFunctionoption. You can select the algorithm with the optionMethod->"LevenbergMarquardt"– Massimo Ortolano May 09 '14 at 09:01NormFunctionoption to the desired minimization function? – dr.blochwave May 09 '14 at 09:02FindMinimumto minimize this directly. The Levenberg-Marquardt method is only applicable to minimands that are explicitly sums of squares, so I would suggest you use the Broyden-Fletcher-Goldfarb-Shanno (BFGS) method instead (Method -> "QuasiNewton"). – Oleksandr R. May 09 '14 at 09:02NormFunctiondocumentation seems to be misleading, because it doesn't correspond directly to the function to be minimized, but actually how the residuals are to be normed. In your case you aren't working with only the residuals, so I don't think this is applicable. – Oleksandr R. May 09 '14 at 09:11FindMinimum- if I wanted to fit e.g. a Gaussian function to noisy data according to the minimization above, how would I go about doing so? (I guess this is difference toNonlinearModelFit, where I can specify a Gaussian with free parameters as the model to be fitted). – dr.blochwave May 09 '14 at 09:15GeneralizedLinearModelFit, which can be used for fitting Poisson-distributed data directly. – Oleksandr R. May 09 '14 at 09:15NormFunction. However, one can probably use an indirect approach by restating the problem so that the given chi-square becomes the model and then can minimize the sum of the model values. – Massimo Ortolano May 09 '14 at 09:22