One thing I use Mathematica for is building molecular graphs.
Until now, I've been content with generating an adjacency matrix from SMILES data, and passing that to AdjacencyGraph. That works well for small cyclic graphs, but the method fails my expectations when the molecules are polycyclic. I want all the edges to be of unit length, but none of the GraphLayout option values permit that.
GraphEdit
The GraphUtilities package contains an interactive graphing function. It enables one to draw any graph, and outputs the relevant data to reconstructing it.
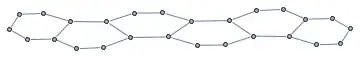
It would be of great help if I could pre-define graphs for GraphEdit to use, that could be copied any number of times inside the applet, while retaining their shapes. This screenshot (from ChemDraw) illustrates what I'm looking for:

Also, directly passing a Graph object to GraphEdit doesn't work:
GraphEdit[CycleGraph[6]]
(* GraphEdit[{1 \[UndirectedEdge] 2, 2 \[UndirectedEdge] 3, 3 \[UndirectedEdge] 4,
4 \[UndirectedEdge] 5, 5 \[UndirectedEdge] 6, 6 \[UndirectedEdge] 1}] *)
Can I add such functionality to GraphEdit?



Head@CycleGraphisGraph,GraphEditdoesn't accept it. – CHM Aug 13 '12 at 04:22GraphEdit. I updated my answer. – Vitaliy Kaurov Aug 13 '12 at 05:35