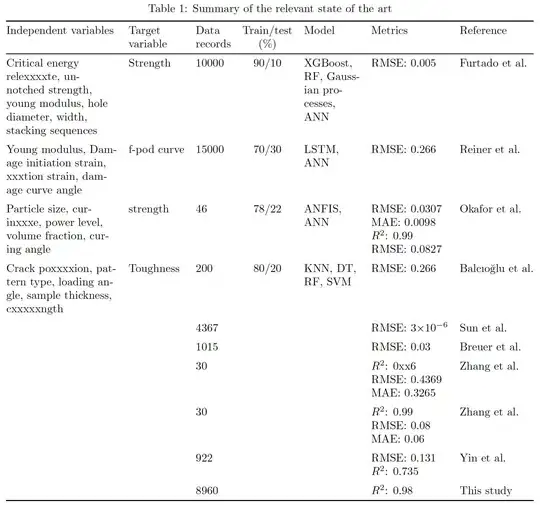
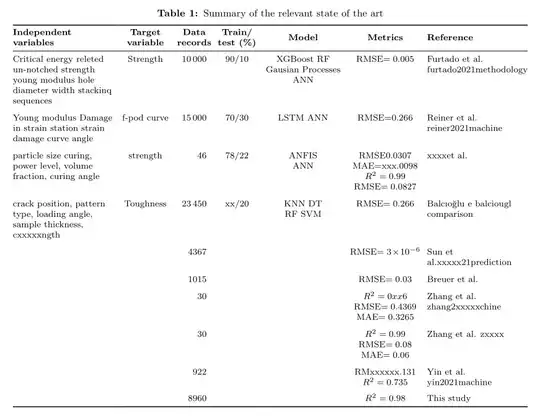
My table width was too much but anyhow I could handle that. Now the length is too much it is going out of margin. What should I do?
My table width was too much but anyhow I could handle that. Now the length is too much it is going out of margin. What should I do?
\documentclass[review]{elsarticle}
%\documentclass[1p]{elsarticle}
\usepackage{geometry}
\usepackage{epsfig}
\usepackage{latexsym}
\usepackage[font={normalsize}]{caption}
\usepackage[para,flushleft]{threeparttable}
\renewcommand{\TPTtagStyle}{\textit}
\usepackage{booktabs}
%\usepackage{nicematrix}
\usepackage{amsmath}
\usepackage{fullpage}
\usepackage{setspace}
\usepackage{mathtools}
\usepackage{fancyhdr}
\usepackage{sectsty}
\usepackage{siunitx}
\usepackage{tabularx}
\usepackage{float}
\usepackage{indentfirst}
\usepackage{changepage}
\usepackage{adjustbox}
\usepackage{tabularx}
\usepackage{rotating}
\usepackage{makecell}
\usepackage{subcaption}
\usepackage{rotating}
%\usepackage{subfigure}
% packages for tables
\usepackage{multirow}
\usepackage[table,xcdraw]{xcolor}
\usepackage{graphicx}
\usepackage{graphics}
\usepackage{caption}
\usepackage{adjustbox}
%\usepackage[normalem]{ulem}
%\usepackage[usenames, dvipsnames]{color}
%\usepackage{nomencl}
%\usepackage{cite}
\usepackage{natbib}
%\usepackage[numbers,super]{natbib}
%\setcitestyle{numbers,super}
\usepackage{enumitem}
\usepackage{lineno,hyperref}
\modulolinenumbers[5]
%\renewcommand{\thefootnote}
\newcommand\degrees[1]{\ensuremath{#1^\circ}}
\newcommand\mycaption[1]{\caption{\footnotesize{#1}}}
\newcommand\myfootnote[1]{\footnote{\small{#1}}}
\newcommand\scalemath[2]{\scalebox{#1}{\mbox{\ensuremath{\displaystyle #2}}}}
\renewcommand{\TPTtagStyle}{\textit}
\newcommand{\splitcell}[1]{\begin{tabular}{@{}c@{}}#1\end{tabular}}
\setlist[enumerate]{label=\arabic.}
\bibliographystyle{elsarticle-num}
\begin{document}
\begin{table}[H]
\centering
%\setlength{\tabcolsep}{2pt} % <--- should be local, anyway
\caption{Summary of the relevant state of the art}
\label{SART}% <--- the label refers to the caption
\begin{tabular}{@{}ccccccl@{}}
\toprule
% headers
\splitcell{Independent \ variables} &
\splitcell{Target \ variable} &
\splitcell{Data \ records} &
Train/test (%) &
Model &
Metrics &
Reference
\
\midrule
% body
\splitcell{Critical energy relexxxxte \ un-notched strength\ young modulus\ hole diameter \ width\ stackixxxxxquences} & Strength & 10000 & 90/10 & \splitcell{XGBoost\RF\Gauxxxxan Processes \ANN}&
RMSE = 0.005 &
Furtado et al. furtado2021methodology
\ \addlinespace
\splitcell{Young modulus\ Damage inxxxxstrain\ xxxtion strain\ damage curve angle} & f-pod curve & 15000 & 70/30 & \splitcell{LSTM\ANN}&
RMSE=0.266 &
Reiner et al. reiner2021machine
\ \addlinespace
\splitcell{particle size\ curinxxxe,power level\ volume fraction\ curing angle} & strength & 46 & 78/22 & \splitcell{ANFIS\ANN}&
\splitcell{RMSE0.0307\ MAE=xxx.0098\ $R^2$=0.99\ RMSE=0.0827} &xxxxet al.
\ \addlinespace
\splitcell{crack poxxxxion\ pattern type\ loading angle\
sample thickness\cxxxxxngth} & Toughness & 2xxx0 & xx/20 & \splitcell{KNN\ DT\ RF\SVM}&
RMSE=0.266 & Balcıoğlu exxxx balciouglxxxxmparison
\ \addlinespace
& & 4367 & & &
RMSE =
3$\times$10$^{-6}$ &
Sun et al.xxxxx21prediction
\ \addlinespace
& & 1015 & & &
RMSE = 0.03 &
Breuer et al.
\ \addlinespace
& & 30 & & &
\splitcell{$R^2$ = 0xx6 \ RMSE = 0.4369 \ MAE = 0.3265} &
Zhang et al. zhang2xxxxxchine
\ \addlinespace
& & 30 & & &
\splitcell{$R^2$ = 0.99 \ RMSE = 0.08 \ MAE = 0.06} &
Zhang et al. zxxxx
\ \addlinespace
& & 922 & & &
\splitcell{RMxxxxxx.131 \ $R^2$ = 0.735} &
Yin et al. yin2021machine
\ \addlinespace
& & 8960 & & &
$R^2$ = 0.98 &
This study
\
\bottomrule
\end{tabular}
\end{table}