Requesting help... Dear all, I have 2 questions if you could please help solve.
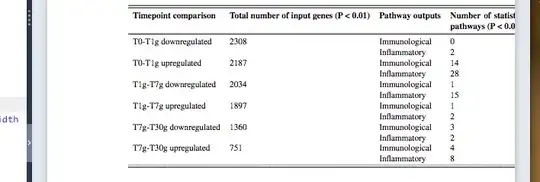
(1) Would anyone please be able to let me know a way to get this table within the margins of the document? Right now, the heading of the second column seems too wide. May be if you can suggest a way that I can shrink the header for the second column down to two lines, it might work. Please see image 1 attached. I have used the following code (please see the code below):
\begin{table}[H]
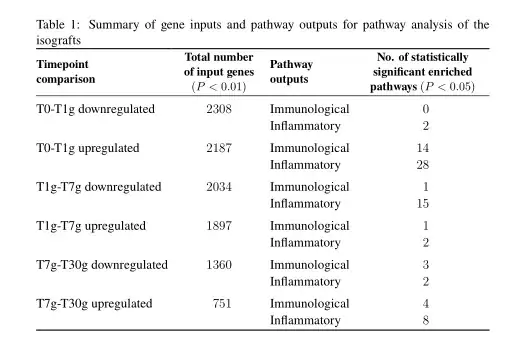
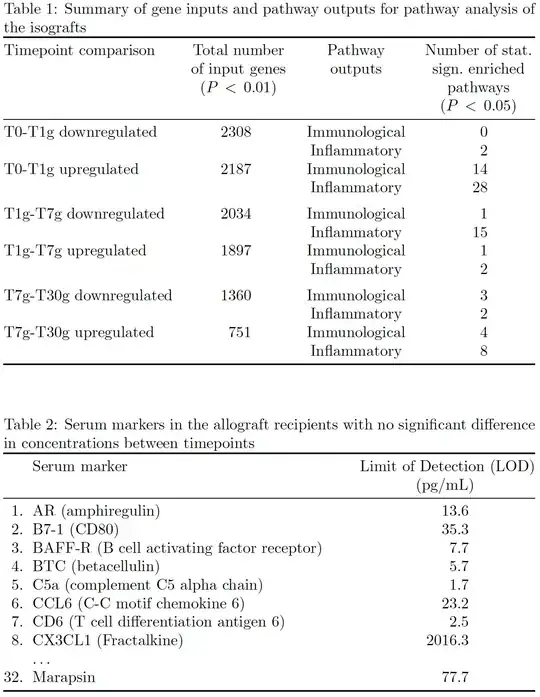
\caption{Summary of gene inputs and pathway outputs for pathway analysis of the isografts}
\small
\centering
\begin{tabular}{lllp{\dimexpr 0.65\linewidth-8\tabcolsep}}
\toprule[\heavyrulewidth]\toprule[\heavyrulewidth]
\textbf{Timepoint comparison} & \textbf{Total number of input genes (P < 0.01)} & \textbf{Pathway outputs} & \textbf{Number of statistically significant enriched pathways (P < 0.05)}\\
\midrule
T0-T1g downregulated & 2308 & Immunological & 0\\
& & Inflammatory & 2 \\
T0-T1g upregulated & 2187 & Immunological & 14 \\
& & Inflammatory & 28 \\
T1g-T7g downregulated & 2034 & Immunological & 1\\
& & Inflammatory & 15 \\
T1g-T7g upregulated & 1897 & Immunological & 1\\
& & Inflammatory & 2 \\
T7g-T30g downregulated & 1360 & Immunological & 3\\
& & Inflammatory & 2 \\
T7g-T30g upregulated & 751 & Immunological & 4\\
& & Inflammatory & 8 \\\hdashline
\bottomrule[\heavyrulewidth]
\end{tabular}
\end{table}
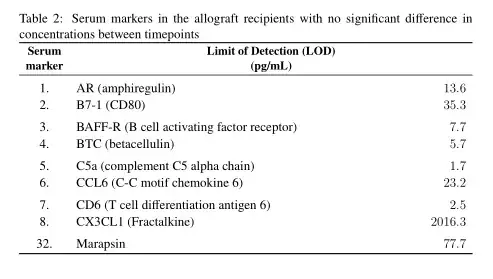
(2) Similar issue as the previous table, except this time, the header of the second column is in two lines, but doesn't align with the rest of the column. Would anyone please be able to let me know a way to align the header with rest of the content in the column? Please see image 2 attached.I have used the following code (please see the code below):
\begin{table}[H]
\setlength{\tabcolsep}{2.5pt}
\caption{Serum markers in the allograft recipients with no significant difference in concentrations between timepoints}
\small
\centering
\begin{tabular}{ l p{\dimexpr 0.65\linewidth-2\tabcolsep}}
\toprule[\heavyrulewidth]\toprule[\heavyrulewidth]
\textbf{Serum marker} & \textbf{\makecell {Limit of Detection (LOD)\\(pg/mL)}}\\
\midrule
1. AR (amphiregulin) & 13.6\\
2. B7-1 (CD80) & 35.3\\
3. BAFF-R (B cell activating factor receptor) & 7.7\\
4. BTC (betacellulin) & 5.7\\
5. C5a (complement C5 alpha chain) & 1.7\\
6. CCL6 (C-C motif chemokine 6) & 23.2\\
7. CD6 (T cell differentiation antigen 6) & 2.5\\
8. CX3CL1 (Fractalkine) & 2016.3\\
32. Marapsin & 77.7\\\hdashline
\bottomrule[\heavyrulewidth]
\end{tabular}
\end{table}
Please see the following details about some parameters I have used for the code as it may help;
\documentclass[12pt, a4paper]{report}
\input{Packages.tex} included the following;
% For tables
\usepackage{threeparttable}
\usepackage{threeparttablex}
\usepackage{ctable}
\usepackage{tabularx}
\usepackage{rotating}
\usepackage{makecell}
\usepackage{longtable}
% other
\usepackage[T1]{fontenc}
\usepackage[latin1]{inputenc}
\usepackage[english]{babel}
\usepackage{siunitx}
\usepackage{graphicx}
\usepackage{tipa} % for the \ark{} command
\usepackage{graphics} % for pdf, bitmapped graphics files
\usepackage{times} % assumes new font selection scheme installed
\usepackage{amsmath}
\usepackage{latexsym}
\usepackage{amscd}% for commutative diagrams
\usepackage{mathrsfs} %this package is for the script font \mathscr
\usepackage{relsize}
\usepackage{delarray}
\usepackage{pstricks}
\usepackage{theorem}
\usepackage{changepage}
\usepackage{euscript}
\usepackage{textcomp}
\usepackage{esvect}
\usepackage{parskip}
\usepackage{placeins}
\usepackage{subfigure}

\theadtpo enable line breaks in the headers. For the second question, about the alignment, use\makecell[l]{…}or\thead[l]since by default it is centred, both horizontally and vertically. Also, note that bydefaul thead fontsize is\footnotesize, so if you do not want it, you have to redefine\theadfont`. – Bernard Mar 28 '22 at 05:36p{\dimexpr 0.65\linewidth-2\tabcolsep}here? You may want to replace it with anlorctype column to get closer to the expected output. – leandriis Mar 28 '22 at 06:24geoemetry. Parameters that you currently did not share. – leandriis Mar 28 '22 at 06:27packages.tex, unfortunately not really. Please also add this information directly into your question using the "edit" button. Code in the comments get overlooked too easily. – leandriis Mar 28 '22 at 06:40threeparttableandctableseem to be incompatible resulting in an error message upon trying to compile your code. Before trying to adjust the output to your liking, make sure the code compiles without any error messages. – leandriis Mar 28 '22 at 06:51booktabsandarydshlnthat define\topruleand\hdashlineare currently missing. – leandriis Mar 28 '22 at 06:53booktabsandarydshln: yes. Adding them to the preamble ys you suggested makes the commands you tried to use available. Regardingthreeparttable/ctable: What to do depends. If you use the commands of one of the packages, keep that package and comment out the other. If you don't use any commands from either package, comment out or remove both of them. – leandriis Mar 28 '22 at 07:02\usepackage[latin1]{inputenc}is correct and needed? Do give\usepackage[utf8]{inputenc}a try. – Mico Mar 28 '22 at 08:00