My generic comment on analysing and identifying patterns in your (pst) documents still holds.
However, here's a way to work backwards through document-refactoring.
1. Your target documentation
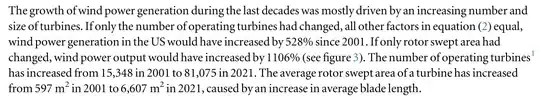
Taking your screenshot your desired target document may look like this in Latex, neglecting all conventions on typesetting numbers, units etc. for demonstration purposes:
\documentclass[10pt,a4paper]{article}
\begin{document}
% ~~~ target documentation ~~~~~~~~~~~
The growth of wind power \dots US would have changed by 528 % since 2001. \dots increased by 1106 % \dots increased from 15,348 in 2001 to 81,075 in 2021. \dots increased from 597 $m^2$ in 2001 to 6,607 $m^2$ in 2021, \dots .
\end{document}
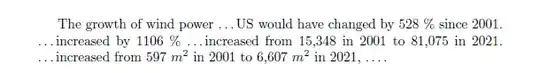
2. Refactoring placeholders
Next let's try to introduce placeholders, which allows you to:
- write text before simulation results become available
- define a set of useful placeholders for that purpose
The complexity of the placeholders content is up to you, and certainly a matter of context. Assuming your simulation also creates some kind of comparision I'd place-out a bit more complex text than just numbers.
Now, the best tool for this is using \newcommand rather than \def. (You could also assign these things to tables, databases etc., it doesn't matter.) Unfortunately your naming convention then is somewhat restricted. However, it might be a good idea to follow a scheme like:
- < dat >< purpose >< differntiator >
- e.g. datCompA
- or simply datA
\documentclass[10pt,a4paper]{article}
\newcommand\datA[0]{528 % since 2001}
\newcommand\datB[0]{1106 %}
\newcommand\datC[0]{15,348 in 2001}
\newcommand\datD[0]{81,075 in 2021}
\newcommand\datE[0]{597 $m^2$ in 2001}
\newcommand\datF[0]{6,607 $m^2$ in 2021}
\begin{document}
% ~~~ refactoring simulated data (placeholders ~~~
The growth of wind power \dots US would have changed by \datA{}. \dots increased by \datB{} \dots increased from \datC{} to \datD{}. \dots increased from \datE{} to \datF{}, \dots .
\end{document}
3. Refactoring for simulation data
Ok, it still compiles as intended. So let's move out those \newcommands:
Target Doc:
\documentclass[10pt,a4paper]{article}
% ~~~ refactoring placeholders ~~~
\input{simdat}
\begin{document}
The growth of wind power \dots US would have changed by \datA{}. \dots increased by \datB{} \dots increased from \datC{} to \datD{}. \dots increased from \datE{} to \datF{}, \dots .
\end{document}
simdat.tex:
\newcommand\datA[0]{528 \% since 2001}
\newcommand\datB[0]{1106 \%}
\newcommand\datC[0]{15,348 in 2001}
\newcommand\datD[0]{81,075 in 2021}
\newcommand\datE[0]{597 $m^2$ in 2001}
\newcommand\datF[0]{6,607 $m^2$ in 2021}
4. Outline of next refactoring steps
Fine. All you now need to do is to generate the content for simdat.tex, e.g. by:
- postprocessing your stored simulation results, using e.g. scripting languages
- creating and storing said content by your simulation program
- etc.
E.g. it should be a no-brainer to go from data here:
528
2001
...
to content there:
\newcommand\datA[0]{528 \% since 2001}
...
5. Conceptual suggestions
For this specific example this procedures seems to be useful:
- prepare your text with required placeholders, best beforehand
- derive required content of placeholders
- create those placeholders from your simulations
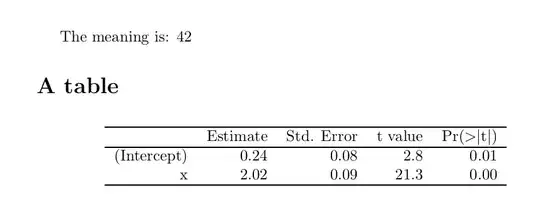
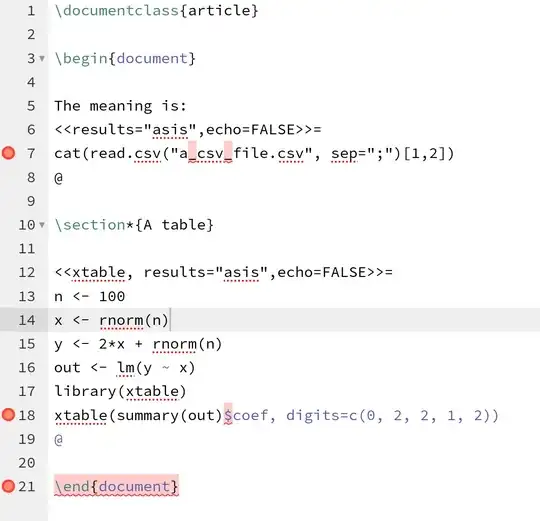
- compile the target doc once simulation runs are done.

\verbatiminput{results.txt}from verbatim or other packages just to show a raw log, or you can use some kind of parser script on the log to make html tables or whatever or of course you can get your original application to log in latex syntax so you can input it directly. All are possible. – David Carlisle Jan 17 '24 at 15:42pgfplotstable, which supports doing this. E.g. see here https://tex.stackexchange.com/search?q=pgfplotstable or here: https://mirror.funkfreundelandshut.de/latex/graphics/pgf/contrib/pgfplots/doc/pgfplotstable.pdf. You may also be tempted to usepgfplotsas well. – MS-SPO Jan 18 '24 at 07:52