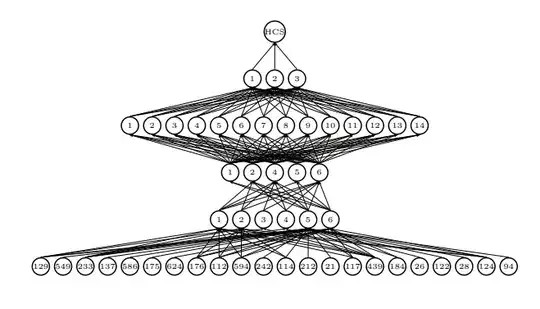
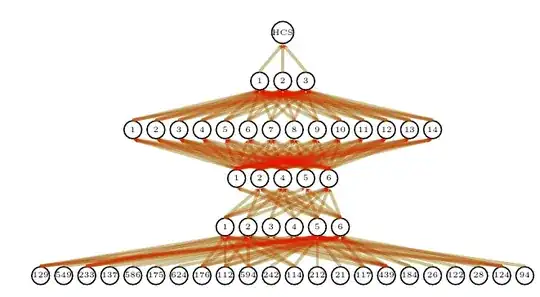
How could the lines be connected to signify density(heat) by the number of lines connecting to parenting nodes. I.e the more lines connect to same parent node, the brighter the heat of the converging line. The lines can be at 90 degree angles, straight or curved, but they should converge somehow. Is there a reasnoable way to represent this data? We can already see the heatmap effect happening on the picture by the density of black connecting lines, however it should be more obvious what node has most connections incoming.
Perhaps making the lines slightly opaque and let stacking create the visual effect of density? Perhaps applying some coloring?
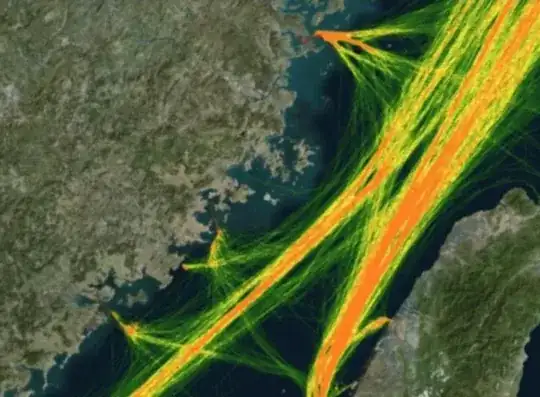
What is density heatmap for the lines? For some inspiration:
\documentclass[12pt, a4paper]{report}
\usepackage{pgfplots}
\usepackage{subcaption}
\usepackage{pgfplots}
\usepackage{xstring}
\usepackage[ruled,vlined]{algorithm2e}
\usepackage{tikz}
\usepackage{amsmath,caption}
\usepackage{svg}
\usepackage{tikz}
\usetikzlibrary{calc,shapes.geometric, arrows, positioning, arrows.meta, fit, backgrounds, patterns, arrows.meta,bending}
\newcommand\drawNodesSuperTight[2]{
\foreach \neurons [count=\lyrIdx] in #2 {
\StrCount{\neurons}{,}[\lyrLength]
\foreach \n [count=\nIdx] in \neurons {
\pgfmathsetmacro\xpos{(\nIdx-1)\nodespacingSuperTight-\nodespacingSuperTight(\lyrLength-1)/2}
\node[neuron2] (#1-\lyrIdx-\nIdx) at (\xpos, \layerdistanceSuperTight*\lyrIdx) {\n};
}
}
}
\newcommand\connectSomeNodesSmallArrow[2]{
\foreach \layer [count=\lyrIdx, evaluate=\lyrIdx as \nextLyr using int(\lyrIdx+1)] in #2
\foreach \neuron [count=\nIdx] in \layer
\foreach \edge in \neuron
\draw[my_arrow2] (#1-\lyrIdx-\nIdx.north) -- (#1-\nextLyr-\edge.south);
}
\newcommand{\nodeFontSuperTight}{\fontsize{4pt}{4pt}\selectfont}
\newcommand{\nodesizeSuperTight}{0.4cm}
\newcommand{\nodepaddingSuperTight}{0.01ex}
\newcommand{\nodespacingSuperTight}{0.1ex}
\newcommand{\layerdistanceSuperTight}{6ex}
\newcommand{\nodesize}{0.5cm}
\newcommand{\nodepadding}{0.2ex}
\newcommand{\nodespacing}{0.3ex}
\newcommand{\nodespacingThight}{0.15ex}
\newcommand{\layerdistance}{8ex}
\tikzset{
my_arrow/.style={-{Stealth[length=3pt]}, line width=0.1mm},
my_arrow2/.style={-{Stealth[length=1.5pt]}, line width=0.1mm},
neuron/.style={circle, draw, font=\tiny, minimum size=\nodesize, inner sep=\nodepadding, thick},
neuron2/.style={circle, draw, font=\nodeFontSuperTight, minimum size=\nodesizeSuperTight, inner sep=\nodepaddingSuperTight, thick},
legend/.style={font=\tiny, fill=white, inner sep=1pt},
odds/.style={legend,fill=white,pos=0.5,inner sep=2pt},
severity/.style={legend,right=1.4ex,align=left,anchor=west},
}
\begin{document}
\begin{tikzpicture}
\drawNodesSuperTight{fcnn}{{{129,549,233,137,586,175,624,176,112,594,242,114,212,21,117,439,184,26,122,28,124,94},{1,2,3,4,5,6},{1,2,4,5,6},{1,2,3,4,5,6,7,8,9,10,11,12,13,14},{1,2,3},{HCS}}}
\connectSomeNodesSmallArrow{fcnn}{{{{1,2,5,6},5,{1,2,3,4,5},5,5,5,6,{1,2,5,6},{1,2,5,6},{1,2,5,6},5,{1,2,5},5,5,{1,5,6},{1,2,3,4,5,6},{5,6},5,5,5,{1,2,5,6},5},{{2,3,4,5},{2,3,5},{3,5},{2,3,5},{1,2,3,4,5},{1,2,3,4,5}},{{1,2,3,4,5,6,7,8,9,10,11,12,13,14},{1,2,3,4,5,6,7,8,9,10,11,12,13,14},{1,2,3,4,5,6,7,8,9,10,11,12,13,14},{3,5,6,7,8,9,10,12,14},{1,2,3,4,5,6,7,8,9,10,11,12,13}},{{1,2,3},{1,2,3},{1,2,3},{1,2,3},{1,2,3},{1,2,3},{1,2,3},{1,2,3},{1,2,3},{1,2,3},{1,2,3},{1,2,3},{1,2,3},{1,2,3}},{1,1,1}}}
\end{tikzpicture}
\end{document}