While the modiagram package does not cover a full range of orbitals, it is still worth using in concert with lower-level TikZ directives for constructing more involved cases. I have used the package in illustrating crystal field theory; for example, the case of a pure sigma donor might look like:
\documentclass{article}
\usepackage{modiagram}
\usepackage{upgreek}
\begin{document}
\begin{figure}
\centering
\begin{MOdiagram}[lines = gray]
\small
% Metal
\AO[metal-3d-1]{s}{-0.10;}
\AO[metal-3d-2]{s}{-0.05;}
\AO[metal-3d-3]{s}{ 0.00;}
\AO[metal-3d-4]{s}{ 0.05;}
\AO[metal-3d-5]{s}{ 0.10;}
% Complex
\AO[complex-t2g-1](50 pt){s}{-1.05;}
\AO[complex-t2g-2](50 pt){s}{-1.00;}
\AO[complex-t2g-3](50 pt){s}{-0.95;}
\AO[complex-eg-1] (50 pt){s}{1.50;}
\AO[complex-eg-2] (50 pt){s}{1.55;}
\node[inner sep = 0, outer sep = 0]
(midway) at (55 pt, 0 pt) {};
\draw[style = dotted] (45 pt, 0 pt) -- ++ (10 pt, 0 pt);
\connect{
metal-3d-3 & complex-t2g-2,
metal-3d-3 & complex-eg-1
}
\node[right] at (complex-t2g-1.east){$\mathrm{t}_{2\mathrm{g}}$};
\node[right] at (complex-eg-1.east) {$\mathrm{e}_{\mathrm{g}}$};
\draw[orange, <->] (complex-t2g-3.west) -- (complex-eg-1.west)
node[midway,left] {$\Delta_{\mathrm{O}}$} ;
\draw[orange, <->] (complex-eg-1.east) -- (midway.east)
node[midway,right] {$\frac{3}{5}\Delta_{\mathrm{O}}$} ;
\draw[orange, <->] (complex-t2g-3.east) -- (midway.east)
node[midway,right] {$\frac{2}{5}\Delta_{\mathrm{O}}$} ;
\end{MOdiagram}
\caption{Octahedral field splitting}
\end{figure}
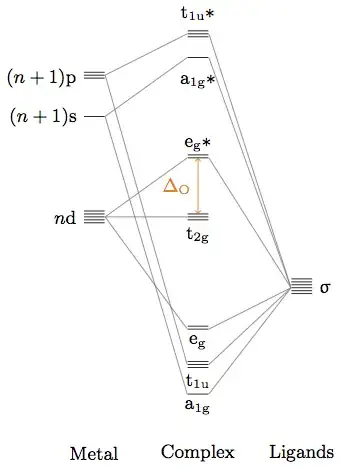
\begin{figure}
\centering
\begin{MOdiagram}[lines= gray]
\small
% Metal
\AO[metal-3d-1]{s}{3.20;}
\AO[metal-3d-2]{s}{3.25;}
\AO[metal-3d-3]{s}{3.30;}
\AO[metal-3d-4]{s}{3.35;}
\AO[metal-3d-5]{s}{3.40;}
\AO[metal-4s] {s}{5.00;}
\AO[metal-4p-1]{s}{5.65;}
\AO[metal-4p-2]{s}{5.70;}
\AO[metal-4p-3]{s}{5.75;}
% Ligand
\AO[ligand-1](100 pt){s}{2.00;}
\AO[ligand-2](100 pt){s}{2.05;}
\AO[ligand-3](100 pt){s}{2.10;}
\AO[ligand-4](100 pt){s}{2.15;}
\AO[ligand-5](100 pt){s}{2.20;}
\AO[ligand-6](100 pt){s}{2.25;}
% Complex
\AO[complex-a1g] (50 pt){s}{0.30;}
\AO[complex-t1u-1] (50 pt){s}{0.75;}
\AO[complex-t1u-2] (50 pt){s}{0.80;}
\AO[complex-t1u-3] (50 pt){s}{0.85;}
\AO[complex-eg-1] (50 pt){s}{1.40;}
\AO[complex-eg-2] (50 pt){s}{1.45;}
\AO[complex-t2g-1] (50 pt){s}{3.25;}
\AO[complex-t2g-2] (50 pt){s}{3.30;}
\AO[complex-t2g-3] (50 pt){s}{3.35;}
\AO[complex-eg*-1] (50 pt){s}{4.30;}
\AO[complex-eg*-2] (50 pt){s}{4.35;}
\AO[complex-a1g*] (50 pt){s}{6.00;}
\AO[complex-t1u*-1](50 pt){s}{6.35;}
\AO[complex-t1u*-2](50 pt){s}{6.40;}
\AO[complex-t1u*-3](50 pt){s}{6.45;}
\connect
{
metal-3d-3 & complex-eg-1 ,
metal-3d-3 & complex-t2g-2 ,
metal-3d-3 & complex-eg*-1 ,
metal-4s & complex-a1g ,
metal-4s & complex-a1g* ,
metal-4p-2 & complex-t1u-2 ,
metal-4p-2 & complex-t1u*-2,
complex-a1g & ligand-3 ,
complex-a1g* & ligand-3 ,
complex-t1u-2 & ligand-3 ,
complex-t1u*-2 & ligand-3 ,
complex-eg-1 & ligand-3 ,
complex-eg*-1 & ligand-3
}
\node[left] at (metal-3d-3.west) {$n\mathrm{d}$};
\node[left] at (metal-4s.west) {$(n + 1)\mathrm{s}$};
\node[left] at (metal-4p-1.west) {$(n + 1)\mathrm{p}$};
\node[below] at (complex-a1g) {$\mathrm{a}_{1\mathrm{g}}$};
\node[below] at (complex-t1u-1) {$\mathrm{t}_{1\mathrm{u}}$};
\node[below] at (complex-eg-1) {$\mathrm{e}_{\mathrm{g}}$};
\node[below] at (complex-t2g-1) {$\mathrm{t}_{2\mathrm{g}}$};
\node[above] at (complex-eg*-1) {$\mathrm{e}_{\mathrm{g}}*$};
\node[below] at (complex-a1g*.south) {$\mathrm{a}_{1\mathrm{g}}*$};
\node[above] at (complex-t1u*-1.north){$\mathrm{t}_{1\mathrm{u}}*$};
\node[right] at (ligand-3.east) {$\upsigma$};
\draw[orange, <->] (complex-t2g-3.center) -- (complex-eg*-1.center)
node[midway,left] {$\Delta_{\mathrm{O}}$} ;
\node at ( 0 pt, -20 pt) {Metal};
\node at ( 50 pt, -20 pt) {Complex};
\node at (100 pt, -20 pt) {Ligands};
\end{MOdiagram}
\caption{Octahedral splitting with a pure $\upsigma$-donor}
\end{figure}
\end{document}

:-)You might want to have a look at our starter guide to familiarize yourself further with our format. Generally, it's best to mark package names with ``` backticks to make them stand out. We also generally avoid using names in posts since it automatically appears in the lower right corner of your post. Welcome! – Adam Liter Nov 25 '13 at 03:26modiagram– cgnieder Nov 25 '13 at 09:27