You cannot use the external tikzlibrary with chemfig because its \tikzexternal@laTeX@collect@until@end@tikzpicture macro, when it executes \begin{tikzpicture}, collects everything until it finds \end{tikzpicture}.
There is an issue when external tikzlibrary is used with chemfig because:
- there is no
\end{tikzpicture} but a \endtikpicture;
- the
\begintikpicture and \endtikpicture do not belong to the
same macro.
This lead to an error. It is possible to fix it by wrapping every \chemfig{<code>} between a \begin{tikzpicture} and \end{tikspicture}, but if you have many \chemfig commands, this requires time.
It is also possible to modify the chemfig code to make it automatically:
\documentclass{article}
\usepackage{chemfig}
\usetikzlibrary{external}
\tikzexternalize
\makeatletter
\def\CF@chemfig@iii[#1][#2]{%
\edef\CF@tmp@str{\noexpand\begin{tikzpicture}[remember picture,every node/.style={anchor=base,inner sep=\z@,outer sep=\z@,minimum size=\z@\ifx\@empty#2\@empty\else,#2\fi},baseline\ifx\@empty#1\@empty\else,#1\fi]}%
\begingroup
\everyeof{\@nil}\endlinechar\m@ne
\CF@sanitize@catcode
\CF@chemfig@iv
}
\def\CF@chemfig@iv#1{%
\CF@tmp@str
\let\CF@hook@list\@empty
\ifx\CF@atom@sep\@empty\def\CF@atom@sep{3em}\fi
\ifx\CF@cram@basewidth\@empty\def\CF@cram@basewidth{1.5ex}\fi
\CF@incyclefalse
\CF@cnt@groupnumber\z@
\let\CF@last@action\z@% 0=d\'ebut du dessin 1=trac\'e d'un noeud 2=trac\'e d'une liaison
\let\CF@start@offset\@empty
\let\CF@end@offset\@empty
\let\CF@bond@outcontentsaved\@empty
\def\CF@cycle@anglecorrection{180/\CF@cycle@num}%
\def\CF@default@angle{0}%
\def\CF@default@stringangle{:0}% angle pris par d\'efaut si le champ est vide
\def\CF@default@length{1}%
\let\CF@default@fromatom\@empty% numero de l'atome d'o\`u partent les liaisons par d\'efaut
\let\CF@default@toatom\@empty% num\'ero de l'atome o\`u arrivent les laisons par d\'efaut
\let\CF@default@tikz\@empty
\let\CF@previous@bondangle\empty
\let\CF@joinbond\z@
\let\CF@previous@tikz\empty
\expandafter\assign@tonil\expandafter\CF@remain@molecule\scantokens{#1}%
\expandafter\CF@chemfig@v\expandafter{\CF@remain@molecule}%
\end{tikzpicture}%
\endgroup
\let\CF@split@state\z@
}
\def\CF@chemfig@vi{%
\let\CF@next@action\CF@chemfig@vi% \`a priori, on reboucle
\ifx\CF@remain@molecule\@empty
\let\CF@next@action\empty
\else
\CF@seek@submol
\expandafter\CF@seek@node\expandafter{\CF@remain@molecule}\CF@current@atomgroup\CF@remain@molecule
\ifx\@empty\CF@current@atomgroup% pas de noeud pour commencer ?
\def\CF@bond@outnode{n\CF@last@groupnumber-%
\ifx\CF@current@fromatom\@empty
\ifdim\CF@current@angle pt<90pt \number\CF@cnt@atomingroup
\else\ifdim\CF@current@angle pt>270pt \number\CF@cnt@atomingroup\else1\fi
\fi
\else\CF@current@fromatom
\fi}%
\expandafter\futurelet\expandafter\CF@toks@a\expandafter\CF@gobble@tonil\CF@remain@molecule\relax\@nil
\CF@if@firsttokin@i{-=<>~}% la suite est une liaison
{\ifnum\CF@last@action=\tw@% c'est la deuxi\`eme liaison cons\'ecutive ?
\CF@insert@emptygroup\CF@remain@molecule% ins\`ere un groupe vide
\edef\CF@bond@outnode{\CF@bond@outnode}%
\else
\ifCF@incycle\advance\CF@cnt@cycle\@ne\fi
\expandafter\CF@analyse@bond\expandafter{\CF@remain@molecule}\CF@bond@type
\edef\CF@bond@outnode{\CF@bond@outnode}%
\let\CF@remain@molecule\CF@remain@afterbond
\ifCF@incycle
\ifnum\CF@cnt@cycle=\CF@cycle@num\relax
\expandafter\expandafter\expandafter\@firstoftwo
\else
\ifnum\CF@cnt@cycle=\@ne
\let\CF@cycle@firsttikz\CF@current@tikz
\unless\ifx\CF@start@offset\@empty\let\CF@cycle@joinlast\z@\fi
\fi
\expandafter\expandafter\expandafter\@secondoftwo
\fi
\else
\expandafter\@secondoftwo
\fi
{\let\CF@next@action\endgroup
\CF@draw@bond\CF@bond@type{\CF@bond@outnode}{\CF@hook@cycle}\CF@previous@atomgroup\CF@hook@atomgroup
}%
{\node[at=(\CF@bond@outnode\ifCF@incycle\else\ifCF@macrofixedbondlength.\CF@current@angle\fi\fi),shift=(\ifcase\CF@split@state\or180-\or-\fi\CF@current@angle:\CF@current@length*\CF@atom@sep)](CF@node){};
\let\CF@previous@angle\CF@current@angle
\let\CF@last@action\tw@
}%
\fi
\ifcat\relax\detokenize\expandafter{\romannumeral-`\.\expandafter\noexpand\CF@remain@molecule}\relax
% s'il ne reste plus rien apr\`es la liaison (sans tenir compte de l'espace devant)-> ins\`ere un groupe vide
\CF@insert@emptygroup\CF@remain@molecule
\fi
}%
{\edef\CF@bond@outnode{\CF@bond@outnode}% \'evalue le l'atome de d\'epart de liaison
\CF@ifx(\CF@toks@a% une parenth\`ese pour commencer ?
{\ifnum\CF@last@action=\tw@% il y avait une liaison juste avant ?
\CF@insert@emptygroup\CF@remain@molecule
\else
\expandafter\CF@grab@submol\expandafter{\CF@remain@molecule}%
\begingroup
\ifCF@incycle\let\CF@last@action\thr@@\fi% on \'etait dans un cycle
\CF@incyclefalse
\aftergroup\CF@chemfig@vi
\def\CF@next@action{\expandafter\CF@chemfig@v\expandafter{\CF@sub@mol}}%
\fi
}%
{\ifx\CF@remain@molecule\@empty
\let\CF@next@action\endgroup
\else% ce qui reste apr\`es le noeud courant n'est pas vide, ne commence pas par "-=~", ni par une parenth\`ese
\CF@ifx*\CF@toks@a% un cycle ?
{\ifnum\CF@last@action=\tw@
\CF@insert@emptygroup\CF@remain@molecule% ins\`ere un groupe vide
\else
\ifCF@incycle\let\CF@last@action\thr@@\fi% on \'etait dans un cycle
\ifnum\CF@last@action=\thr@@\let\CF@lastcycle@num\CF@cycle@num\fi
\CF@expafterarg@ntimes2\CF@if@firsttokmatch{\expandafter\@gobble\CF@remain@molecule}*% un 2\`e "*" ?
{\CF@expafterarg@ntimes2{\def\CF@remain@molecule}{\expandafter\@gobble\CF@remain@molecule}% enl\`eve la 1er \'etoile
\CF@expafterarg@ntimes2\CF@if@firsttokmatch{\expandafter\@gobble\CF@remain@molecule}[% un crochet ensuite ?
{\expandafter\CF@cycle@parsepreamblewithoptarg\CF@remain@molecule\@nil% \begingroup inclus
}%
{\def\CF@cyclearc@startangle{0}\def\CF@cyclearc@endangle{360}%
\let\CF@cyclearc@directtikz\@empty
\expandafter\CF@cycle@parsepreamble\CF@remain@molecule\@nil% \begingroup inclus
}%
\CF@cyclearctrue
}%
{\expandafter\CF@cycle@parsepreamble\CF@remain@molecule\@nil% \begingroup inclus
\CF@cyclearcfalse
}%
\CF@cnt@cycle\z@
\edef\CF@hook@cycle{\CF@bond@outnode}%
\let\CF@hook@atomgroup\CF@previous@atomgroup
\ifx\CF@hook@atomgroup\CF@empty@node
\let\CF@cycle@joinlast\@ne% joindre le dernier
\else
\let\CF@cycle@joinlast\z@
\fi
\CF@incycletrue
\ifnum\CF@last@action=\thr@@
\pgfmathparse{360/\CF@lastcycle@num-180}% c'est un cycle dans un cycle
\else
\pgfmathparse{-180/\CF@cycle@num-90+\CF@cycle@anglecorrection}%
\fi
\let\CF@initcycle@angle\pgfmathresult
\aftergroup\CF@chemfig@vi
\def\CF@next@action{\expandafter\CF@chemfig@v\expandafter{\CF@sub@mol}}%
\fi
}%
{\errmessage{Package \CF@package@name\space Error: something went wrong here: \detokenize\expandafter{\CF@remain@molecule}^^JIf you think it's a bug, please, send a Minimal Example to the author.}}%
\fi}%
}%
\else
\CF@expand@second{\CF@expand@second{\CF@expand@second\CF@draw@atomgroup\CF@current@angle}\CF@current@toatom}\CF@current@atomgroup
\fi
\fi
\CF@next@action
}
\makeatother
\begin{document}
\chemname{\chemfig{A=B-C-D}}{example}
\end{document}
You can check that the image of the molecule is recorded in a pdf file (<name of master file>-image0.pdf) in the current directory. Here is the output:

There is a small issue though... Since the bounding box of a C is smaller than its outline, the cropping done by external is too tight:
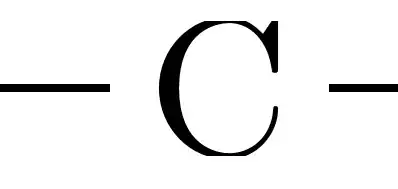
To fix that, you can increse the inner sep and set it to 1pt instead of \z@ (which is 0pt):
\edef\CF@tmp@str{\noexpand\begin{tikzpicture}[remember picture,every node/.style={anchor=base,inner sep=1pt,outer sep=\z@,minimum size=\z@\ifx\@empty#2\@empty\else,#2\fi},baseline\ifx\@empty#1\@empty\else,#1\fi]}%

chemfigis based on tikz, and the standard way to avoid unnecessary recompilations in tikz is to use either\usetikzlibrary{external}or thestandalonepackage. I would expect that both work (although I am unfamiliar withchemfig). Theexternallib should work as soon as you write\usetikzlibrary{external}\tikzexternalizeinto the preamble and compile withpdflatex -shell-escape .... Thestandalonepackage requires more work, but might result in a better reusable molecule database. If you need more details, you will have to edit your question to show us what you have. – Christian Feuersänger May 14 '15 at 21:37