I have quite some experience with R, however none with LaTeX. So any help would be appreciated. I have a data frame in R and used this code line to export it into the console:
print(xtable(plot_data),floating=FALSE,latex.environments=NULL,booktabs=TRUE)
which leads to the following output:
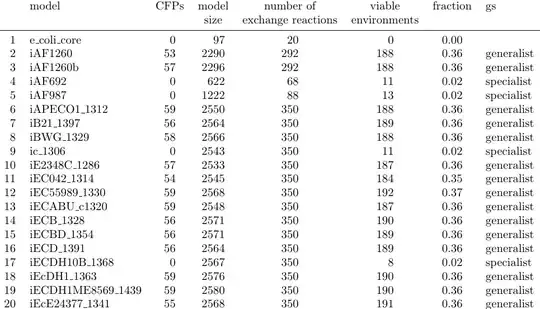
\begin{tabular}{rlrrrrrl}
\toprule
& model & CFPs & model\_size & number\_of\_exchange\_reactions & viable\_environments & fraction & gs \\
\midrule
1 & e\_coli\_core & 0 & 97 & 20 & 0 & 0.00 & \\
2 & iAF1260 & 53 & 2290 & 292 & 188 & 0.36 & generalist \\
3 & iAF1260b & 57 & 2296 & 292 & 188 & 0.36 & generalist \\
4 & iAF692 & 0 & 622 & 68 & 11 & 0.02 & specialist \\
5 & iAF987 & 0 & 1222 & 88 & 13 & 0.02 & specialist \\
6 & iAPECO1\_1312 & 59 & 2550 & 350 & 188 & 0.36 & generalist \\
7 & iB21\_1397 & 56 & 2564 & 350 & 189 & 0.36 & generalist \\
8 & iBWG\_1329 & 58 & 2566 & 350 & 188 & 0.36 & generalist \\
9 & ic\_1306 & 0 & 2543 & 350 & 11 & 0.02 & specialist \\
10 & iE2348C\_1286 & 57 & 2533 & 350 & 187 & 0.36 & generalist \\
11 & iEC042\_1314 & 54 & 2545 & 350 & 184 & 0.35 & generalist \\
12 & iEC55989\_1330 & 59 & 2568 & 350 & 192 & 0.37 & generalist \\
13 & iECABU\_c1320 & 59 & 2548 & 350 & 187 & 0.36 & generalist \\
14 & iECB\_1328 & 56 & 2571 & 350 & 190 & 0.36 & generalist \\
15 & iECBD\_1354 & 56 & 2571 & 350 & 189 & 0.36 & generalist \\
16 & iECD\_1391 & 56 & 2564 & 350 & 189 & 0.36 & generalist \\
17 & iECDH10B\_1368 & 0 & 2567 & 350 & 8 & 0.02 & specialist \\
18 & iEcDH1\_1363 & 59 & 2576 & 350 & 190 & 0.36 & generalist \\
19 & iECDH1ME8569\_1439 & 59 & 2580 & 350 & 190 & 0.36 & generalist \\
20 & iEcE24377\_1341 & 55 & 2568 & 350 & 191 & 0.36 & generalist \\
21 & iECED1\_1282 & 57 & 2537 & 350 & 181 & 0.34 & generalist \\
22 & iECH74115\_1262 & 53 & 2533 & 350 & 181 & 0.34 & generalist \\
23 & iEcHS\_1320 & 55 & 2562 & 350 & 189 & 0.36 & generalist \\
24 & iECIAI1\_1343 & 51 & 2552 & 344 & 185 & 0.35 & generalist \\
25 & iECIAI39\_1322 & 51 & 2532 & 350 & 184 & 0.35 & generalist \\
26 & iECNA114\_1301 & 57 & 2550 & 350 & 190 & 0.36 & generalist \\
27 & iECO103\_1326 & 57 & 2573 & 350 & 192 & 0.37 & generalist \\
28 & iECO111\_1330 & 57 & 2573 & 350 & 190 & 0.36 & generalist \\
29 & iECO26\_1355 & 57 & 2592 & 350 & 193 & 0.37 & generalist \\
30 & iECOK1\_1307 & 59 & 2544 & 350 & 186 & 0.35 & generalist \\
31 & iEcolC\_1368 & 54 & 2581 & 350 & 191 & 0.36 & generalist \\
32 & iECP\_1309 & 59 & 2557 & 350 & 192 & 0.37 & generalist \\
33 & iECs\_1301 & 54 & 2555 & 350 & 189 & 0.36 & generalist \\
34 & iECS88\_1305 & 57 & 2544 & 350 & 187 & 0.36 & generalist \\
35 & iECSE\_1348 & 57 & 2581 & 350 & 193 & 0.37 & generalist \\
36 & iECSF\_1327 & 59 & 2567 & 350 & 190 & 0.36 & generalist \\
37 & iEcSMS35\_1347 & 59 & 2562 & 350 & 190 & 0.36 & generalist \\
38 & iECSP\_1301 & 54 & 2546 & 350 & 189 & 0.36 & generalist \\
39 & iECUMN\_1333 & 57 & 2568 & 350 & 191 & 0.36 & generalist \\
40 & iECW\_1372 & 56 & 2585 & 350 & 192 & 0.37 & generalist \\
41 & iEKO11\_1354 & 56 & 2583 & 350 & 192 & 0.37 & generalist \\
42 & iETEC\_1333 & 57 & 2565 & 350 & 190 & 0.36 & generalist \\
43 & iG2583\_1286 & 54 & 2537 & 350 & 185 & 0.35 & generalist \\
44 & iHN637 & 13 & 759 & 94 & 57 & 0.11 & specialist \\
45 & iIT341 & 0 & 529 & 73 & 6 & 0.01 & specialist \\
46 & iJN678 & 8 & 712 & 47 & 34 & 0.06 & specialist \\
47 & iJN746 & 14 & 1012 & 87 & 89 & 0.17 & specialist \\
48 & iJO1366 & 58 & 2451 & 307 & 191 & 0.36 & generalist \\
49 & iJR904 & 32 & 1049 & 140 & 141 & 0.27 & generalist \\
50 & iLF82\_1304 & 60 & 2542 & 350 & 186 & 0.35 & generalist \\
51 & iLJ478 & 0 & 606 & 74 & 0 & 0.00 & \\
52 & iMM904 & 31 & 1331 & 162 & 114 & 0.22 & generalist \\
53 & iND750 & 22 & 1076 & 115 & 91 & 0.17 & specialist \\
54 & iNJ661 & 16 & 984 & 84 & 78 & 0.15 & specialist \\
55 & iNRG857\_1313 & 60 & 2549 & 350 & 186 & 0.35 & generalist \\
56 & iPC815 & 0 & 1875 & 274 & 8 & 0.02 & specialist \\
57 & iRC1080 & 0 & 1404 & 18 & 18 & 0.03 & specialist \\
58 & iS\_1188 & 43 & 2456 & 350 & 172 & 0.33 & generalist \\
59 & iSB619 & 6 & 675 & 83 & 8 & 0.02 & specialist \\
60 & iSbBS512\_1146 & 43 & 2430 & 350 & 148 & 0.28 & generalist \\
61 & iSBO\_1134 & 39 & 2436 & 350 & 158 & 0.30 & generalist \\
62 & iSDY\_1059 & 43 & 2382 & 350 & 147 & 0.28 & generalist \\
63 & iSF\_1195 & 0 & 2468 & 350 & 9 & 0.02 & specialist \\
64 & iSFV\_1184 & 41 & 2457 & 350 & 167 & 0.32 & generalist \\
65 & iSFxv\_1172 & 45 & 2475 & 350 & 168 & 0.32 & generalist \\
66 & iSSON\_1240 & 47 & 2521 & 350 & 175 & 0.33 & generalist \\
67 & iUMN146\_1321 & 59 & 2550 & 350 & 186 & 0.35 & generalist \\
68 & iUMNK88\_1353 & 57 & 2581 & 350 & 191 & 0.36 & generalist \\
69 & iUTI89\_1310 & 59 & 2541 & 350 & 186 & 0.35 & generalist \\
70 & iWFL\_1372 & 56 & 2585 & 350 & 192 & 0.37 & generalist \\
71 & iY75\_1357 & 59 & 2584 & 350 & 190 & 0.36 & generalist \\
72 & iYL1228 & 29 & 2146 & 272 & 157 & 0.30 & generalist \\
73 & iYO844 & 34 & 1062 & 186 & 191 & 0.36 & generalist \\
74 & iZ\_1308 & 54 & 2556 & 350 & 189 & 0.36 & generalist \\
75 & STM\_v1\_0 & 46 & 2407 & 326 & 170 & 0.32 & generalist \\
76 & iSM199 & 0 & 278 & 36 & 0 & 0.00 & \\
\bottomrule
\end{tabular}
Now, here comes the hard part. I know, that this has to be imported into LaTeX. What lines of code do I have to add to make it work and to make the table formatted in a right way (such that I can directly use it as a .pdf or .png)?
I have downloaded TeXmaker and can also use an online version of LaTeX with this


Sweaveand its sucessorknitr. There are many example of knitr in this site. For example here. – Fran Jul 13 '19 at 15:03