\documentclass[12pt]{elsarticle}
%% for removing the footer
\makeatletter
\def\ps@pprintTitle{%
\let\@oddhead\@empty
\let\@evenhead\@empty
%\let\@oddfoot\@empty
\let\@evenfoot\@oddfoot
}
\usepackage{multirow}
\usepackage{lineno} %% package for line number
%% The amssymb package provides various useful mathematical symbols
\usepackage{lscape}
\usepackage{graphicx}
\usepackage{subfig}
\usepackage{multicol}
\setlength{\columnsep}{1cm}
\usepackage[utf8]{inputenc}
\usepackage[T1]{fontenc}
\usepackage{textcomp}
\usepackage{amssymb}
\usepackage{url}
\usepackage{showframe}
\renewcommand\ShowFrameLinethickness{0.15pt}
\renewcommand*\ShowFrameColor{\color{red}}
\usepackage{afterpage,lscape}
\usepackage{lipsum, pdflscape}
\usepackage[noabbrev,nameinlink]{cleveref}
\usepackage[margin=25mm]{geometry}
\usepackage{graphicx}
\usepackage{tabularray}
\UseTblrLibrary{booktabs}
\begin{document}
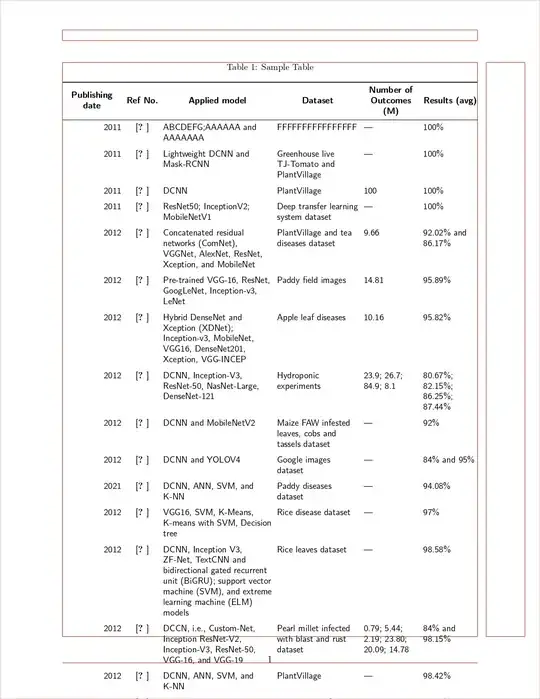
\begin{table}
\caption{Sample Table}
\footnotesize\sffamily
\begin{tblr}{colsep= 3pt,
colspec={@{}r c X[2, l] X[1.5,l] X[l] X[l] @{}},
row{1} = {font=\bfseries, c, m},
rowsep= 3pt
}
\toprule
\textbf {Publishing \\date} & \textbf {Ref No.} & \textbf {Applied model} & \textbf {Dataset} & \textbf {Number of Outcomes (M)} & \textbf {Results (avg)} \\
\midrule
2011 & \cite{--------} & ABCDEFG;AAAAAA and AAAAAAA & FFFFFFFFFFFFFFFF & --- & 100\% \\
2011 & \cite{-------------} & Lightweight DCNN and Mask-RCNN & Greenhouse live TJ-Tomato and PlantVillage & --- & 100\% \\
2011 & \cite{---------} & DCNN & PlantVillage & 100 & 100\% \\
2011 & \cite{----------} & ResNet50; InceptionV2; MobileNetV1 & Deep transfer learning system dataset & --- & 100\% \\
2012 & \cite{-----------} & Concatenated residual networks (ComNet), VGGNet, AlexNet, ResNet, Xception, and MobileNet & PlantVillage and tea diseases dataset & 9.66 & 92.02\% and 86.17\% \\
2012 & \cite{-----------} & Pre-trained VGG-16, ResNet, GoogLeNet, Inception-v3, LeNet & Paddy field images & 14.81 & 95.89\% \\
2012 & \cite{-------------} & Hybrid DenseNet and Xception (XDNet); Inception-v3, MobileNet, VGG16, DenseNet201, Xception, VGG-INCEP & Apple leaf diseases & 10.16 & 95.82\% \\
2012 & \cite{--------} & DCNN, Inception-V3, ResNet-50, NasNet-Large, DenseNet-121 & Hydroponic experiments & 23.9; 26.7; 84.9; 8.1 & 80.67\%; 82.15\%; 86.25\%; 87.44\% \\
2012 & \cite{---------} & DCNN and MobileNetV2 & Maize FAW infested leaves, cobs and tassels dataset & --- & 92\% \\
2012 & \cite{---------} & DCNN and YOLOV4 & Google images dataset & --- & 84\% and 95\%\\
2021 & \cite{------------} & DCNN, ANN, SVM, and K-NN & Paddy diseases dataset & --- & 94.08\% \\
2012 & \cite{------------} & VGG16, SVM, K-Means, K-means with SVM, Decision tree & Rice disease dataset & --- & 97\% \\
2012 & \cite{-------} & DCNN, Inception V3, ZF-Net, TextCNN and bidirectional gated recurrent unit (BiGRU); support vector machine (SVM), and extreme learning machine (ELM) models & Rice leaves dataset & --- & 98.58\% \\
2012 & \cite{----------} & DCCN, i.e., Custom-Net, Inception ResNet-V2, Inception-V3, ResNet-50, VGG-16, and VGG-19 & Pearl millet infected with blast and rust dataset & 0.79; 5.44; 2.19; 23.80; 20.09; 14.78 & 84\% and 98.15\% \\
2012 & \cite{----------} & DCNN, ANN, SVM, and K-NN & PlantVillage & --- & 98.42\% \\
2012 & \cite{-----------} & AlexNet, SqueezeNet, GoogLeNet, ResNet-50, and ResNet-101 & Pearl millet infected with blast and Guava disease dataset & --- & 96.01\% \\
2012 & \cite{-----------} & AlexNet, GoogleNet, VGG16, and VGG19 & Healthy and infected maize dataset & --- & 99.94\% \\
2012 & \cite{-----------} & DCNN, SVM, Decision tree, Linear Regression, K-NN, AlexNet, ResNet, VGG16, InceptionV3 & Alternaria Leaf Spot (ALS) disease dataset & --- & 98.41\% \\
2012 & \cite{---------} & YOLO-V5, VGG-16, AlexNet, ResNet-50, ResNet-101, DenseNet-121 and Bidirectional Cross-Modal Transformer (BiCMT) & Xiaotangshn National Precision Agriculture Demonstration Base dataset & --- & 96.92\% \\
2012 & \cite{--------} & YEYEYEYE, VGG16, VGG19, MobileNetV2, DHDHDHHDD, ResNet-V2, NasNetMobile, Inception-V3 and InceptionResNetV2 & Tipburn Disorder Detection in Strawberry Leaves dataset & 1.008; ;134.27; 139.578; 0.23; 7.77; 21.81; 58.34; 54.34; 4.27 & 96.03\% \\
\bottomrule
\end{tblr}
\end{table}
\end{document}
- 23
- 5
1 Answers
Welcome to TeX.SE!
Try to make columns with short text in cell make narrower and column with text in several lines, wider. By use of the tabularray package this can be easily done:
\documentclass{article}
\usepackage[margin=25mm]{geometry}
%---------------- show page layout. don't use in a real document!
\usepackage{showframe}
\renewcommand\ShowFrameLinethickness{0.15pt}
\renewcommand*\ShowFrameColor{\color{red}}
%---------------------------------------------------------------%
\usepackage{graphicx}
\usepackage{tabularray}
\UseTblrLibrary{booktabs}
\begin{document}
\begin{table}
\caption{My caption}
\label{my-label}
\footnotesize\sffamily
\begin{tblr}{colsep= 4pt,
colspec={@{}r c X[2, l] X[1.5,l] X[l] X[l] @{}},
row{1} = {font=\bfseries, c, m},
rowsep=3pt
}
\toprule
Yea & SL No. & DL model & Dataset & {No. of\ parameter (M)} & {Accuracy (Avg)} \
\midrule
1000 & \cite{1} & Matrix-based CNN; AlexNet and VGG-16 & Winter wheat leaf diseases images & --- & 93.1% \
2000 & \cite{2} & Lightweight DCNN and Mask-RCNN & Greenhouse live TJ-Tomato and PlantVillage & --- & 93.61% \
3000 & \cite{3} & DCNN & PlantVillage & 0.766 & 93.03% \
4000 & \cite{4} & ResNet50; InceptionV2; MobileNetV1 & Deep transfer learning system dataset & --- & 91% \
5020 & \cite{5} & Concatenated residual networks (ComNet), VGGNet, AlexNet, ResNet, Xception, and MobileNet & PlantVillage and tea diseases dataset & 98.10 & 90.41% and 86.17% \
20000 & \cite{6} & Pre-trained VGG-16, ResNet, GoogLeNet, Inception-v3, LeNet & Paddy field images & 14.81 & 93.13% \
1111 & \cite{7} & Hybrid DenseNet and Xception (XDNet); Inception-v3, MobileNet, VGG16, DenseNet201, Xception, VGG-INCEP & Apple leaf diseases & 10.16 & 91.02% \
5643 & \cite{8} & DCNN, Inception-V3, ResNet-50, NasNet-Large, DenseNet-121 & Hydroponic experiments & 23.9; 26.7; 84.4; 8.1 & 41.67%; 95.15%; 76.25%; 27.44% \
8356 & \cite{9} & DCNN and MobileNetV2 & Maize FAW infested leaves, cobs and tassels dataset & --- & 97% \
8989 & \cite{10} & DCNN and YOLOV4 & Google images dataset & --- & 94% and 45% \
4312 & \cite{11} & DCNN, ANN, SVM, and K-NN & Paddy diseases dataset & --- & 94.08% \
7865 & \cite{12} & VGG16, SVM, K-Means, K-means with SVM, Decision tree & Rice disease dataset & --- & 92% \
7878 & \cite{13} & DCNN, Inception V3, ZF-Net, TextCNN and bidirectional gated recurrent unit (BiGRU); support vector machine (SVM), and extreme learning machine (ELM) models & Rice leaves dataset & --- & 92.58% \
4321 & \cite{14} & DCCN, i.e., Custom-Net, Inception ResNet-V2, Inception-V3, ResNet-50, VGG-16, and VGG-19 & Pearl millet infected with blast and rust dataset & 0.79; 5.44; 2.19; 23.80; 20.09; 14.78 & 84% and 98.15% \
4444 & \cite{15} & DCNN, ANN, SVM, and K-NN & PlantVillage & --- & 91.42% \
6543 & \cite{16} & AlexNet, SqueezeNet, GoogLeNet, ResNet-50, and ResNet-101 & Pearl millet infected with blast and Guava disease dataset & --- & 96.74% \
3456 & \cite{17} & AlexNet, GoogleNet, VGG16, and VGG19 & Healthy and infected maize dataset & --- & 96.94% \
3432 & \cite{18} & DCNN, SVM, Decision tree, Linear Regression, K-NN, AlexNet, ResNet, VGG16, InceptionV3 & Alternaria Leaf Spot (ALS) disease dataset & --- & 98.41% \
9743 & \cite{19} & YOLO-V5, VGG-16, AlexNet, ResNet-50, ResNet-101, DenseNet-121 and Bidirectional Cross-Modal Transformer (BiCMT) & Xiaotangshn National Precision Agriculture Demonstration Base dataset & --- & 96.92% \
3432 & \cite{19} & PSO-CNN, VGG16, VGG19, MobileNetV2, EfficientNet, ResNet-V2, NasNetMobile, Inception-V3 and InceptionResNetV2 & Tipburn Disorder Detection in Strawberry Leaves dataset & 1.008; ;134.27; 139.578; 0.23; 7.77; 21.81; 58.34; 54.34; 6.27 & 67.73% \
\bottomrule
\end{tblr}
\end{table}
\end{document}
Compilation result obtained in Overleaf:
(red lines indicate page layout)
Addendum:
Converting to long table:
\documentclass{article}
\usepackage[margin=25mm]{geometry}
%---------------- show page layout. don't use in a real document!
\usepackage{showframe}
\renewcommand\ShowFrameLinethickness{0.15pt}
\renewcommand*\ShowFrameColor{\color{red}}
%---------------------------------------------------------------%
\usepackage{graphicx}
\usepackage{tabularray}
\UseTblrLibrary{booktabs}
\begin{document}
\begingroup
\small\sffamily\linespread{0.92}\selectfont
\begin{longtblr}[
caption = {My caption},
label = {my-label}
]{rowhead = 1,
colsep= 4pt,
colspec={@{}r c X[2, l] X[1.5,l] X[l] X[l] @{}},
row{1} = {font=\bfseries, c, m},
rowsep=2pt
}
\toprule
Yea &{SL\ No.}& {DL\ model} & Dataset & {No. of\ parameter (M)} & {Accuracy\ (Avg)} \
\midrule
%
% table body is the same as before
%
- 296,517


\documentclass{article}gives the same result as is shown in my answer. I will check you MWE immediately when it will be available. – Zarko Nov 04 '22 at 13:53[p](so far I didn't add it). Sory, my crystal ball is out in order, so I can't see your document ;-). Please edit your question and provide complete small document with some dummy text (generate with ˙lipsum` or similar package) which reproduce your problem. This may give a chance anyone that will be able to help you. – Zarko Nov 14 '22 at 13:4610ptbut your document you use12pt. This can be compensated by reducing font size in table, from\footnotesizeto\scriptsize. But this will make it worse readable. I don't know, if this is acceptable to you. BTW, you cannot put an elephant in lady suitcase :-(. You should consider to split table over two pages by usinglongtblrtable. – Zarko Nov 15 '22 at 09:34