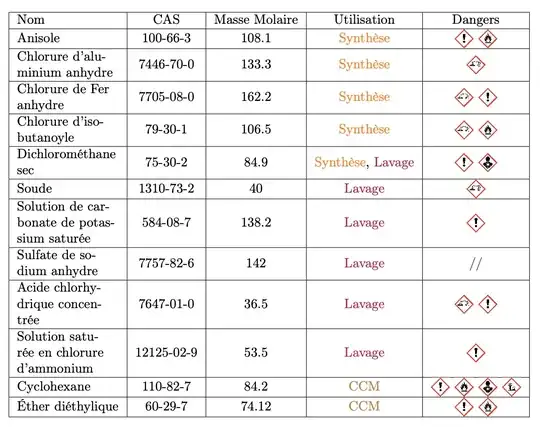
I've run into a problem that I can't seem to find a solution to. I am using the package tabularx since I work with small pages. My table has a mix of figures and text, and I would like the column to be vertically aligned.
Here if you check the 3rd line or the 8th line all the columns are not vertically aligned. Do you have some option to vertically align them please?
\documentclass[10pt]{article}
\usepackage[paperheight=9.45in,paperwidth=6.69in,margin=0.5in,heightrounded]{geometry}
\synctex=1
\usepackage[utf8]{inputenc}
\usepackage[T1]{fontenc}
\usepackage[french]{babel}
\usepackage{xcolor}
\usepackage{tabularx,booktabs}
\usepackage{ghsystem}
\renewcommand\tabularxcolumn[1]{m{#1}}% for vertical centering text in X column
\begin{document}
\begin{center}
\begin{tabularx}{0.95\textwidth}{|X|c|c|c|c|}
\hline
Nom & CAS & Masse Molaire & Utilisation & Dangers \\
\hline
Anisole & 100-66-3 & 108.1 & {\color{orange} Synthèse} & \ghspic[scale=0.4]{exclam} \ghspic[scale=0.4]{flame}\\
\hline
Chlorure d’aluminium anhydre & 7446-70-0 & 133.3 & {\color{orange} Synthèse} & \ghspic[scale=0.4]{acid} \\
\hline
Chlorure de Fer anhydre & 7705-08-0 & 162.2 & {\color{orange} Synthèse} & \ghspic[scale=0.4]{acid} \ghspic[scale=0.4]{exclam}\\
\hline
Chlorure d'isobutanoyle & 79-30-1 & 106.5 & {\color{orange} Synthèse} & \ghspic[scale=0.4]{acid} \ghspic[scale=0.4]{flame} \\
\hline
Dichlorométhane sec & 75-30-2 & 84.9 & {\color{orange} Synthèse}, {\color{purple} Lavage} & \ghspic[scale=0.4]{exclam} \ghspic[scale=0.4]{health} \\
\hline
Soude & 1310-73-2 & 40 & {\color{purple} Lavage} & \ghspic[scale=0.4]{acid} \\
\hline
Solution de carbonate de potassium saturée & 584-08-7 & 138.2 & {\color{purple} Lavage} & \ghspic[scale=0.4]{exclam} \\
\hline
Sulfate de sodium anhydre & 7757-82-6 & 142 & {\color{purple} Lavage} & // \\
\hline
Acide chlorhydrique concentrée & 7647-01-0 & 36.5 & {\color{purple} Lavage} & \ghspic[scale=0.4]{acid} \ghspic[scale=0.4]{exclam} \\
\hline
Solution saturée en chlorure d'ammonium & 12125-02-9 & 53.5 & {\color{purple} Lavage} & \ghspic[scale=0.4]{exclam}\\
\hline
Cyclohexane & 110-82-7 & 84.2 & {\color{brown} CCM} & \ghspic[scale=0.4]{exclam} \ghspic[scale=0.4]{flame} \ghspic[scale=0.4]{health} \ghspic[scale=0.4]{aqpol} \\
\hline
Éther diéthylique & 60-29-7 & 74.12 & {\color{brown} CCM} & \ghspic[scale=0.4]{exclam} \ghspic[scale=0.4]{flame} \\
\hline
\end{tabularx}
\end{center}
\end{document}
Here is the result:


\renewcommand\tabularxcolumn[1]{m{#1}}in the preamble.) – Mico Mar 15 '23 at 22:36