There is one of my "hackish" solutions, not to be taken too seriously.
I'll show first the result, then the high-level code, then the ugly macro which draws one cylinder and its wrapping, and finally the explanations about how I did it all.
1. The result
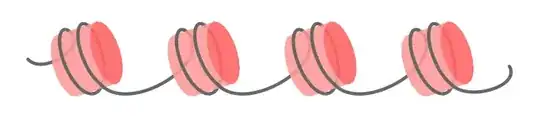
2. High level tikzpicture
It contains a loop in which a strand is drawn from previous nucleosome to the current one, then the nucleosome is drawn on top, and finally some coordinate points are set up for the next iteration.
% Define styles, colors and opacity
\tikzset{
wrapping/.style = {draw=black!60, line cap=round, line join=round, ultra thick},
nucleosome/.style = {red!40, fill opacity=.9, draw=none},
top cylinder/.style= {red!60, fill opacity=.9}
}
\begin{tikzpicture} % Draw the chain
\coordinate (previous) at (-1,0) {};
\coordinate (start) at (-.235,.423) {};
\foreach \x in {0,2,4,6} { % Draw four nucleosomes, 2cm apart each other
\begin{scope}[xshift=\x cm]
\draw[wrapping] (previous) to[out=-50, in=-60] (start); % connection from previous
\nucleosome % Black magic here
\coordinate (previous) at (end) {}; % update join points
\coordinate (start) at ([xshift=2cm] start) {};
\end{scope}
}
% Last strand
\draw[wrapping] (previous) to[out=-50, in=-60] +(1,.3);
\end{tikzpicture}
3. \nucleosome macro
Be warned, it is ugly. After it, the explanation of where the numbers came from:
\def\nucleosome{
\draw[wrapping](.198,-.549)--(.204,-.543)--(.225,-.519)--(.243,-.49)--(.26,-.457)--(.274,-.42)--(.285,-.379)--(.294,-.335)--(.3,-.288)--(.303,-.238)--(.304,-.186)--(.302,-.133)--(.297,-.078)--(.29,-.023)--(.281,.033)--(.27,.089)--(.256,.143)--(.24,.197)--(.223,.249)--(.204,.299)--(.184,.347)--(.163,.391)--(.14,.432)--(.117,.47)--(.094,.504)--(.071,.533)--(.047,.558)--(.024,.578)--(.002,.593)--(-.012,.599);
\draw[wrapping](-.235,.423)--(-.259,.449)--(-.282,.471)--(-.305,.488)--(-.325,.499);
%\filldraw[fill=green](-.235,.423) circle (2pt);
\coordinate (start) at (-.235,.423) {};
\filldraw[top cylinder](.422,-.414)--(.499,-.422)--(.561,-.379)--(.6,-.288)--(.614,-.159)--(.6,-.003)--(.561,.163)--(.499,.324)--(.422,.464)--(.336,.569)--(.251,.629)--(.173,.638)--(.112,.595)--(.072,.504)--(.059,.374)--(.072,.218)--(.112,.052)--(.173,-.109)--(.251,-.249)--(.336,-.354)--cycle;
\filldraw[nucleosome](-.561,.379)--(.112,.595)--(.173,.638)--(-.499,.422)--cycle;
\draw[wrapping](-.325,.499)--(-.327,.5)--(-.348,.507)--(-.368,.509)--(-.386,.505)--(-.391,.503);
\filldraw[nucleosome](-.173,-.638)--(.499,-.422)--(.422,-.414)--(-.251,-.629)--cycle;
\draw[wrapping](.112,-.59)--(.129,-.587)--(.156,-.578)--(.181,-.563)--(.198,-.549);
\draw[wrapping](-.012,.599)--(-.02,.603)--(-.041,.609)--(-.06,.609)--(-.077,.604);
\filldraw[nucleosome](-.6,.288)--(.072,.504)--(.112,.595)--(-.561,.379)--cycle;
\draw[wrapping](-.391,.503)--(-.403,.497)--(-.418,.484)--(-.431,.466)--(-.441,.443)--(-.45,.415)--(-.454,.392);
\filldraw[nucleosome](-.251,-.629)--(.422,-.414)--(.336,-.354)--(-.336,-.569)--cycle;
\draw[wrapping](-.059,-.538)--(-.057,-.54)--(-.024,-.56)--(.009,-.575)--(.04,-.586)--(.071,-.592)--(.101,-.592)--(.112,-.59);
\draw[wrapping](-.077,.604)--(-.078,.604)--(-.094,.594)--(-.108,.579)--(-.121,.559)--(-.131,.535)--(-.138,.506)--(-.14,.493);
\filldraw[nucleosome](-.614,.159)--(.059,.374)--(.072,.504)--(-.6,.288)--cycle;
\filldraw[nucleosome](-.336,-.569)--(.336,-.354)--(.251,-.249)--(-.422,-.464)--cycle;
\draw[wrapping](-.454,.392)--(-.456,.384)--(-.46,.348)--(-.461,.309)--(-.459,.267)--(-.459,.264);
\filldraw[nucleosome](-.6,.003)--(.072,.218)--(.059,.374)--(-.614,.159)--cycle;
\filldraw[nucleosome](-.422,-.464)--(.251,-.249)--(.173,-.109)--(-.499,-.324)--cycle;
\draw[wrapping](-.167,-.438)--(-.155,-.452)--(-.122,-.486)--(-.089,-.515)--(-.059,-.538);
\filldraw[nucleosome](-.561,-.163)--(.112,.052)--(.072,.218)--(-.6,.003)--cycle;
\filldraw[nucleosome](-.499,-.324)--(.173,-.109)--(.112,.052)--(-.561,-.163)--cycle;
\draw[wrapping](-.459,.264)--(-.455,.222)--(-.448,.175)--(-.438,.125)--(-.433,.105);
\draw[wrapping](-.14,.493)--(-.144,.473)--(-.146,.436)--(-.147,.396)--(-.145,.365);
\draw[wrapping](-.269,-.298)--(-.248,-.331)--(-.218,-.375)--(-.187,-.415)--(-.167,-.438);
\draw[wrapping](-.433,.105)--(-.426,.074)--(-.411,.022)--(-.394,-.03)--(-.38,-.069);
\draw[wrapping](.146,-.337)--(.17,-.363)--(.202,-.395)--(.235,-.423);
%\filldraw[fill=green](.235,-.423) circle (2pt);
\coordinate (end) at (.235,-.423) {};
\draw[wrapping](-.355,-.131)--(-.353,-.135)--(-.329,-.187)--(-.304,-.237)--(-.277,-.285)--(-.269,-.298);
\draw[wrapping](-.38,-.069)--(-.375,-.083)--(-.355,-.131);
\draw[wrapping](-.145,.365)--(-.144,.353)--(-.139,.307)--(-.131,.259)--(-.121,.209)--(-.12,.205);
\draw[wrapping](.044,-.197)--(.047,-.2)--(.076,-.246)--(.106,-.288)--(.138,-.328)--(.146,-.337);
\draw[wrapping](-.12,.205)--(-.108,.158)--(-.092,.105)--(-.074,.053)--(-.066,.032);
\draw[wrapping](-.04,-.031)--(-.032,-.052)--(-.007,-.103)--(.019,-.153)--(.044,-.197);
\draw[wrapping](-.066,.032)--(-.054,0)--(-.04,-.031);
}
4. The explanations
First of all, inspired by @cmhughes comment, I wrote a sketch program to draw a single nucleosome, with part of a helix around it. I marked also with a big green dot the start and end of the helix. These will be the "connecting points" from one nucleosome to the next one. This is the sketch code, note that I use line style=wrapping or fill style=nucleosome to be able to customize the style later from tikz.
def start_angle 180 % starting and ending angles of the helix
def end_angle 720-180
def radius_helix 1.08 % helix radius, a bit more than the cylinder
def cylinder_height 1.4
def percentage_wrapped 70 % How many of the cylinder is wrapped in the helix
def start_point (radius_helix*cos(start_angle),
-percentage_wrapped/100*cylinder_height/2,
radius_helix*sin(start_angle))
def helix {
def n_segs 100
sweep [line style=wrapping]
{ n_segs, translate([0, cylinder_height*percentage_wrapped/100/n_segs,0]),
rotate(end_angle/n_segs, [0,1,0]) } (start_point)
}
def start { % Starting point of the helix
dots [fill=green](start_point)
}
def end { % Ending point of the helix
put { translate([0, cylinder_height*percentage_wrapped/100,0])
then rotate(end_angle, [0,1,0]) } {{start}}
}
def cylinder {
def n_cyl_segs 20
sweep[fill style=top cylinder]{ n_cyl_segs<>,
rotate(360/n_cyl_segs, [0,1,0]) }
line[fill style=nucleosome](1,-cylinder_height/2)(1,cylinder_height/2)
}
def nucleosome {
put { rotate (-60, [1, 0, 0])} {{cylinder}{helix}{start}{end}}
}
put { view((10,15,0)) } { {nucleosome} } % Draw it!
global { language tikz }
I processed the above file with the command:
sketch nucleosome.sketch > nucleosome.tex
Then included the result in the document:
\documentclass{article}
\usepackage{tikz}
\tikzset{
wrapping/.style = {draw=black!60, line cap=round, line join=round, ultra thick},
nucleosome/.style = {red!40, fill opacity=.9, draw=none},
top cylinder/.style= {red!60, fill opacity=.9}
}
\begin{document}
\input nucleosome
\end{document}
And got (please, note the green big dots):

Then edited nucleosome.tex and searched for the string green. I found:
[...]
\filldraw[fill=green](-.235,.423) circle (2pt);
[...]
\filldraw[fill=green](.235,-.423) circle (2pt);
[...]
This gave me the coordinates in the 2D projection of the starting and ending point of the nucleosome wrapping. I manually replaced those lines with:
[...]
%\filldraw[fill=green](-.235,.423) circle (2pt);
\coordinate (start) at (-.235,.423) {};
[...]
%\filldraw[fill=green](.235,-.423) circle (2pt);
\coordinate (end) at (.235,-.423) {};
[...]
Then replaced the \begin{tikzpicture} with \def\nucleosome{ and the \end{tikzpicture} with } to get the definition of the \nucleosome macro, already shown in section 3. The rest should be clear.





 (edit: fixed handedness of wrapping and added background layer)
(edit: fixed handedness of wrapping and added background layer)
PSTrickswill be the way forward; it's hard to do the hidden parts intikz– cmhughes Dec 05 '12 at 04:28