Dataset Processing (for Life Sciences)
Note: a related, but distinct task is posted here ID Swapping: Efficient use of a reference table to convert ID values.
A common task, at least for me, involves analyzing at least two different Datasets.
A common example, at least currently in biological sciences, involve gene lists. Depending on where they came from, who processed them, which pipeline they were processed with, etc they might vary in form.
For example, consider the following two Datasets:
In Dataset One (d1) we see that there are two conditions c1 and c2 with various replicates, e.g. {c1_1, c1_2, c1_3}. In Dataset Two (d2) we do not have the issue of replicates. However, in both sets we have duplicate ids, with varying values in their columns. Biologically this might arise if one converted the transcript id (a subset of the gene) to the gene id. Lastly, not all genes are in both sets. Therefore we have some preprocessing to do:
- find those IDs common to both sets
- combining into a singular
Dataset - find duplicates rows (e.g. same value in the ID column)
- average those duplicates rows by column value
- replace those duplicates with their average
- find replicates in the column headers
- average those columns together
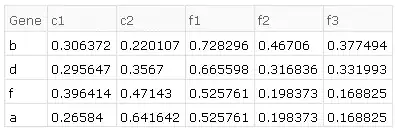
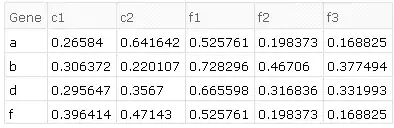
i.e. applying the above transformations to given datasets, we should end up with:
There are a lot of ways to approach this problem. Below I am including what I made to answer this post. However I am sure it is not the most efficient (or most elegantly coded) method. Thus I would appreciate your assistance in finding better ways at this kind of pre-processing.
Shout out to @OneSquare 's answer on Variable named slots.
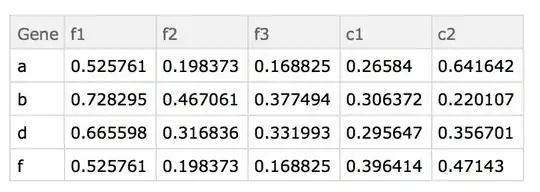
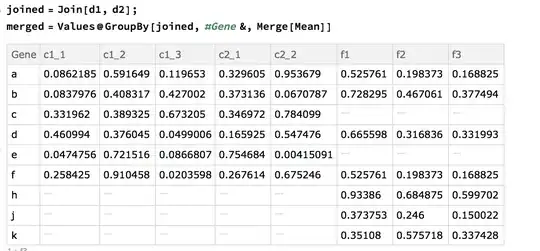
Here are the "datasets" used:
d1=Dataset@{<|"Gene" -> "a", "c1_1" -> 0.0862185, "c1_2" -> 0.591649,
"c1_3" -> 0.119653, "c2_1" -> 0.329605,
"c2_2" -> 0.953679|>, <|"Gene" -> "b", "c1_1" -> 0.0837976,
"c1_2" -> 0.408317, "c1_3" -> 0.427002, "c2_1" -> 0.373136,
"c2_2" -> 0.0670787|>, <|"Gene" -> "c", "c1_1" -> 0.331962,
"c1_2" -> 0.389325, "c1_3" -> 0.673205, "c2_1" -> 0.346972,
"c2_2" -> 0.784099|>, <|"Gene" -> "d", "c1_1" -> 0.460994,
"c1_2" -> 0.376045, "c1_3" -> 0.0499006, "c2_1" -> 0.165925,
"c2_2" -> 0.547476|>, <|"Gene" -> "e", "c1_1" -> 0.0474756,
"c1_2" -> 0.721516, "c1_3" -> 0.0866807, "c2_1" -> 0.754684,
"c2_2" -> 0.00415091|>, <|"Gene" -> "f", "c1_1" -> 0.258425,
"c1_2" -> 0.910458, "c1_3" -> 0.0203598, "c2_1" -> 0.267614,
"c2_2" -> 0.675246|>, <|"Gene" -> "c", "c1_1" -> 0.331962,
"c1_2" -> 0.389325, "c1_3" -> 0.673205, "c2_1" -> 0.346972,
"c2_2" -> 0.784099|>, <|"Gene" -> "d", "c1_1" -> 0.460994,
"c1_2" -> 0.376045, "c1_3" -> 0.0499006, "c2_1" -> 0.165925,
"c2_2" -> 0.547476|>, <|"Gene" -> "c", "c1_1" -> 0.331962,
"c1_2" -> 0.389325, "c1_3" -> 0.673205, "c2_1" -> 0.346972,
"c2_2" -> 0.784099|>, <|"Gene" -> "c", "c1_1" -> 0.331962,
"c1_2" -> 0.389325, "c1_3" -> 0.673205, "c2_1" -> 0.346972,
"c2_2" -> 0.784099|>, <|"Gene" -> "a", "c1_1" -> 0.0862185,
"c1_2" -> 0.591649, "c1_3" -> 0.119653, "c2_1" -> 0.329605,
"c2_2" -> 0.953679|>}
d2=Dataset@{<|"Gene" -> "h", "f1" -> 0.93386, "f2" -> 0.684875,
"f3" -> 0.599702|>, <|"Gene" -> "b", "f1" -> 0.93083,
"f2" -> 0.735748, "f3" -> 0.586162|>, <|"Gene" -> "j",
"f1" -> 0.373753, "f2" -> 0.246,
"f3" -> 0.150022|>, <|"Gene" -> "d", "f1" -> 0.945271,
"f2" -> 0.553761, "f3" -> 0.658329|>, <|"Gene" -> "k",
"f1" -> 0.35108, "f2" -> 0.575718,
"f3" -> 0.337428|>, <|"Gene" -> "f", "f1" -> 0.525761,
"f2" -> 0.198373, "f3" -> 0.168825|>, <|"Gene" -> "d",
"f1" -> 0.525761, "f2" -> 0.198373,
"f3" -> 0.168825|>, <|"Gene" -> "d", "f1" -> 0.525761,
"f2" -> 0.198373, "f3" -> 0.168825|>, <|"Gene" -> "a",
"f1" -> 0.525761, "f2" -> 0.198373,
"f3" -> 0.168825|>, <|"Gene" -> "b", "f1" -> 0.525761,
"f2" -> 0.198373, "f3" -> 0.168825|>}
@Kuba's approach (prior to this update), is certainly more succinct and the syntax is a bit foreign to me. It does merge the data sets together and take the mean; however, it does not handle duplicate IDs. The replicate part of this question was added during the update so naturally it was not included in his answer.
Desired exact result
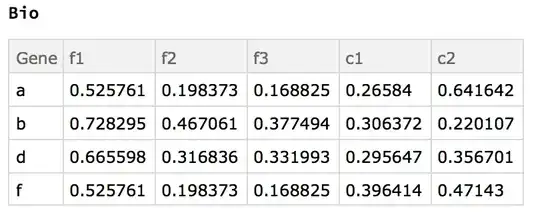
The desired results on the given example data in order of transformations is as follows.
- common ids of both sets:
{"a", "b", "d", "f"} - combining (tack on
d2to the end ofd1 - find duplicate rows, e.g. in
d1there are two rows with the id "a", four with "c", etc - average those duplicates together by id, e.g. for rows with the id "a", looking at only column "c1_1", then the average would be $(0.0862184+0.0862184)/2$.
- find replicates in the column header (e.g. "c1_1", "c1_2", "c1_3" are replicates of "c1")
- average them together, so for "a" and replicates of "c1", $(0.0862184+0.591649+0.119653)/3$.
e.g. produces the result of the example given above













JoinAcrossthat makes sense to me. I was wondering where this function may be useful. Up to now I considered it a retarded sister ofGroupBy+Merge. :) +1 – Kuba Oct 13 '16 at 06:35JoinAcross. My own answer, which appears to be an excessively verbose equivalent to yours, works with an arbitrary number ofDatasetobjects.JoinAcrossrequires the first two arguments be separate lists. if you had a variabled={d1,d2,...}how could you alterJoinAcrossto handle that? I feel like this is a need forFoldbut I never been able to getFoldto work as I wanted to. Also could you possibly elaborate on your use of composition/*? – SumNeuron Oct 21 '16 at 05:44Fold[JoinAcross[##, "Gene"] &, {d1, d2, d3}]. Often it is the case that the join critieria are not identical or there are key collisions between the datasets. In such cases we need to explicitly nest theJoinAcrossexpressions./*essentially chains operators together so that they are applied in order. In queries, the use can be subtle -- see (98193) for discussion. – WReach Oct 21 '16 at 14:56Datasetobjects? Why are some methods unable to work withDatasetif it is just an association wrapped with a different head? – SumNeuron Oct 27 '16 at 06:14KeyValueMapin the operator form ofQueryas follows:data[All, KeyValueMap[...]/*Merge[Mean]], it works. Could you possibly break down that function though? I get the string replace patterns. What I do not understand is why 1.) you use a delayed assignment, 2.) why you wrap the string replace in an association and then use a pure function. It comes to reason that#2is the value associated with the givenKeycorrect? By why this notation? Also where to read more aboutinner? – SumNeuron Oct 27 '16 at 09:40##references all arguments, in this case it is shorthand for#, #2. I don't use delayed assignment -- perhaps you mean:>? I use it instead of->to ensure thatnis a local variable. The notation<| ... |> &is simply defining a pure function that returns an association. The term "inner" here comes from relational joins. I'm happy to continue discussion, but perhaps it should be in chat. – WReach Oct 28 '16 at 03:39JoinAcrossis seriously broken in version 11.0.0 but works just fine in 11.0.1 and 10.x releases -- see the comments to (129122). – WReach Oct 28 '16 at 03:48