Gene List Pre-Processing
ABOUT:
In this example we have two gene lists, both from mice. One gene list contains columns of features, the other has condition replicates.
GOAL:
The goal is to
- Convert the Ensembl IDs to gene names
- Replaces the Ensembl IDs in our Datasets with gene names
- Find those genes common to both Datasets and use only those records
- Find duplicate gene names and average those rows together
- Find replicates in our columns and merge those together
- Combine these datasets into a single Dataset for easier handling
NOTE:
This example uses the code described and demonstrated here:
The example data (because it is "clunky") is at the bottom of this post. An image of it is posted here for convenience.

Get our genes
geneList =
Normal@With[{slot = First@Normal@Keys@First@geneListFile},
geneListFile[All, Slot[slot] &]];
geneList2 =
Normal@With[{slot = First@Normal@Keys@First@geneListFile2},
geneListFile2[All, Slot[slot] &]];

Set up for ID conversion
speciesCommonName = "mouse";
speciesCommonNameAssociations = <|"HUMAN" -> "hg", "MOUSE" -> "mm",
"YEAST" -> "sc", "RAT" -> "rn"|>;
speciesEncodedName =
Lookup[ToUpperCase@speciesCommonName]@
speciesCommonNameAssociations;
referenceFileName =
First@FileNames[___ ~~ speciesEncodedName ~~ ___ ~~ ".csv",
NotebookDirectory[]];
Load reference file
referenceFile = SemanticImport@referenceFileName;
keys = Normal@Keys@First@referenceFile;
data = referenceFile[All,
Select[keys, StringContainsQ[#, "Gene"] &]];
\[ScriptCapitalA] =
AssociationThread[#[[;; , 1]], #[[;; , 2]]] &@Normal@data[Values];
Convert Ensembl IDs to gene names
genes = Lookup[\[ScriptCapitalA], #] & /@ geneList;
genes2 = Lookup[\[ScriptCapitalA], #] & /@ geneList2;

Replace Ensembl ID with gene name
d1 = Dataset@
MapThread[
Prepend, { Normal@geneListFile, Thread["Genes" -> genes]}];
d2 = Dataset@
MapThread[
Prepend, { Normal@geneListFile2, Thread["Genes" -> genes2]}];
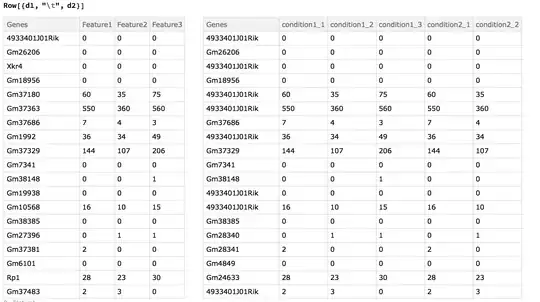
Functions for merging data
DealWithDuplicates[data_] :=
Module[{keys, dupicateValues, duplicatePositions,
duplicatesAveraged}, keys = Normal@Keys@First@data;
dupicateValues =
If[Length[#] > 1, First@#, Nothing] & /@
Split@Normal@data[All, #[First@keys] &];
duplicatePositions =
Flatten[#] & /@ (Position[Normal@data[All, #[First@keys] &], #] & /@
dupicateValues);
duplicatesAveraged =
data[duplicatePositions[[#]]][Mean] & /@
Range[Length@duplicatePositions];
Return[{duplicatePositions, duplicatesAveraged}]]
ReplaceDuplicatesWithMean[data_, duplicatePositions_,
duplicateAveraged_] := Module[{temp}, temp = data;
Table[temp =
ReplacePart[
temp, {First@duplicatePositions[[i]]} ->
Normal@duplicateAveraged[[i]]], {i, Length@duplicateAveraged}];
Return[temp];]
DeleteDuplicatesNotAveraged[data_, duplicatePositions_,
duplicateAveraged_] := Module[{minus, temp}, temp = data;
temp = Delete[temp,
duplicatePositions[[#, 2 ;;]] & /@
Range@Length@duplicatePositions];
Return[temp]]
mergeData[Data_List] :=
Module[{keys, data, common, dupData}, data = Data;
keys = Normal@Keys@First@data[[#]] & /@ Range@Length@data;
data = Table[
With[{key = keys[[d]]}, data[[d]][SortBy[#[First@key] &]]], {d,
Length@data}];
common =
Intersection[
Table[With[{key = keys[[d]]}, data[[d]][All, #[First@key] &]], {d,
Length@data}]];
data = Table[
With[{key = keys[[d]]},
data[[d]][Select[MemberQ[common[[d]], #[First@key]] &]]], {d,
Length@data}];
dupData = Table[DealWithDuplicates[data[[d]]], {d, Length@data}];
data = Table[
ReplaceDuplicatesWithMean[data[[d]], dupData[[d]][[1]],
dupData[[d]][[2]]], {d, Length@data}];
data = DeleteDuplicatesBy[#, First] & /@ data;
Return[JoinAcross @@ Append[data, First@First@keys]]]
Merge Data
Bio = mergeData[{d1, d2}]
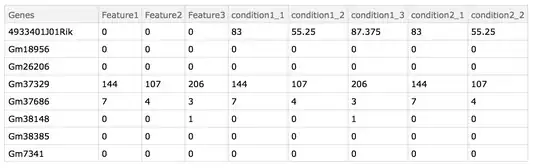
bioKeys = Normal@Keys@First@Bio
conditions = {"condition1", "condition2"};
replicates =
Flatten@Position[bioKeys, #] & /@
Flatten@StringCases[bioKeys, conditions[[#]] ~~ __] & /@
Range@Length@conditions;
mergedReplicates = Bio[All, Flatten[bioKeys[[#]] & /@ replicates[[#]], 1] /* <|
conditions[[#]] -> Mean|>] & /@ Range@Length@conditions;
Bio = Bio[All, Delete[Partition[Flatten@replicates, 1]]];
Table[Bio = Dataset@MapThread[
Append, {Normal@Bio, Thread[Normal@mergedReplicates[[i]]]}], {i,Length@mergedReplicates}];

Example Data
geneListFile=Dataset@{<|"Genes" -> "ENSMUSG00000102693", "Feature1" -> 0, "Feature2" -> 0,
"Feature3" -> 0|>, <|"Genes" -> "ENSMUSG00000064842",
"Feature1" -> 0, "Feature2" -> 0,
"Feature3" -> 0|>, <|"Genes" -> "ENSMUSG00000051951",
"Feature1" -> 0, "Feature2" -> 0,
"Feature3" -> 0|>, <|"Genes" -> "ENSMUSG00000102851",
"Feature1" -> 0, "Feature2" -> 0,
"Feature3" -> 0|>, <|"Genes" -> "ENSMUSG00000103377",
"Feature1" -> 60, "Feature2" -> 35,
"Feature3" -> 75|>, <|"Genes" -> "ENSMUSG00000104017",
"Feature1" -> 550, "Feature2" -> 360,
"Feature3" -> 560|>, <|"Genes" -> "ENSMUSG00000103025",
"Feature1" -> 7, "Feature2" -> 4,
"Feature3" -> 3|>, <|"Genes" -> "ENSMUSG00000089699",
"Feature1" -> 36, "Feature2" -> 34,
"Feature3" -> 49|>, <|"Genes" -> "ENSMUSG00000103201",
"Feature1" -> 144, "Feature2" -> 107,
"Feature3" -> 206|>, <|"Genes" -> "ENSMUSG00000103147",
"Feature1" -> 0, "Feature2" -> 0,
"Feature3" -> 0|>, <|"Genes" -> "ENSMUSG00000103161",
"Feature1" -> 0, "Feature2" -> 0,
"Feature3" -> 1|>, <|"Genes" -> "ENSMUSG00000102331",
"Feature1" -> 0, "Feature2" -> 0,
"Feature3" -> 0|>, <|"Genes" -> "ENSMUSG00000102348",
"Feature1" -> 16, "Feature2" -> 10,
"Feature3" -> 15|>, <|"Genes" -> "ENSMUSG00000102592",
"Feature1" -> 0, "Feature2" -> 0,
"Feature3" -> 0|>, <|"Genes" -> "ENSMUSG00000088333",
"Feature1" -> 0, "Feature2" -> 1,
"Feature3" -> 1|>, <|"Genes" -> "ENSMUSG00000102343",
"Feature1" -> 2, "Feature2" -> 0,
"Feature3" -> 0|>, <|"Genes" -> "ENSMUSG00000102948",
"Feature1" -> 0, "Feature2" -> 0,
"Feature3" -> 0|>, <|"Genes" -> "ENSMUSG00000025900",
"Feature1" -> 28, "Feature2" -> 23,
"Feature3" -> 30|>, <|"Genes" -> "ENSMUSG00000104123",
"Feature1" -> 2, "Feature2" -> 3, "Feature3" -> 0|>}
geneListFile2=Dataset@{<|"Genes" -> "ENSMUSG00000102693", "condition1_1" -> 0,
"condition1_2" -> 0, "condition1_3" -> 0, "condition2_1" -> 0,
"condition2_2" -> 0|>, <|"Genes" -> "ENSMUSG00000064842",
"condition1_1" -> 0, "condition1_2" -> 0, "condition1_3" -> 0,
"condition2_1" -> 0,
"condition2_2" -> 0|>, <|"Genes" -> "ENSMUSG00000102693",
"condition1_1" -> 0, "condition1_2" -> 0, "condition1_3" -> 0,
"condition2_1" -> 0,
"condition2_2" -> 0|>, <|"Genes" -> "ENSMUSG00000102851",
"condition1_1" -> 0, "condition1_2" -> 0, "condition1_3" -> 0,
"condition2_1" -> 0,
"condition2_2" -> 0|>, <|"Genes" -> "ENSMUSG00000102693",
"condition1_1" -> 60, "condition1_2" -> 35, "condition1_3" -> 75,
"condition2_1" -> 60,
"condition2_2" -> 35|>, <|"Genes" -> "ENSMUSG00000102693",
"condition1_1" -> 550, "condition1_2" -> 360, "condition1_3" -> 560,
"condition2_1" -> 550,
"condition2_2" -> 360|>, <|"Genes" -> "ENSMUSG00000103025",
"condition1_1" -> 7, "condition1_2" -> 4, "condition1_3" -> 3,
"condition2_1" -> 7,
"condition2_2" -> 4|>, <|"Genes" -> "ENSMUSG00000102693",
"condition1_1" -> 36, "condition1_2" -> 34, "condition1_3" -> 49,
"condition2_1" -> 36,
"condition2_2" -> 34|>, <|"Genes" -> "ENSMUSG00000103201",
"condition1_1" -> 144, "condition1_2" -> 107, "condition1_3" -> 206,
"condition2_1" -> 144,
"condition2_2" -> 107|>, <|"Genes" -> "ENSMUSG00000103147",
"condition1_1" -> 0, "condition1_2" -> 0, "condition1_3" -> 0,
"condition2_1" -> 0,
"condition2_2" -> 0|>, <|"Genes" -> "ENSMUSG00000103161",
"condition1_1" -> 0, "condition1_2" -> 0, "condition1_3" -> 1,
"condition2_1" -> 0,
"condition2_2" -> 0|>, <|"Genes" -> "ENSMUSG00000102693",
"condition1_1" -> 0, "condition1_2" -> 0, "condition1_3" -> 0,
"condition2_1" -> 0,
"condition2_2" -> 0|>, <|"Genes" -> "ENSMUSG00000102693",
"condition1_1" -> 16, "condition1_2" -> 10, "condition1_3" -> 15,
"condition2_1" -> 16,
"condition2_2" -> 10|>, <|"Genes" -> "ENSMUSG00000102592",
"condition1_1" -> 0, "condition1_2" -> 0, "condition1_3" -> 0,
"condition2_1" -> 0,
"condition2_2" -> 0|>, <|"Genes" -> "ENSMUSG00000100538",
"condition1_1" -> 0, "condition1_2" -> 1, "condition1_3" -> 1,
"condition2_1" -> 0,
"condition2_2" -> 1|>, <|"Genes" -> "ENSMUSG00000101117",
"condition1_1" -> 2, "condition1_2" -> 0, "condition1_3" -> 0,
"condition2_1" -> 2,
"condition2_2" -> 0|>, <|"Genes" -> "ENSMUSG00000100204",
"condition1_1" -> 0, "condition1_2" -> 0, "condition1_3" -> 0,
"condition2_1" -> 0,
"condition2_2" -> 0|>, <|"Genes" -> "ENSMUSG00000084668",
"condition1_1" -> 28, "condition1_2" -> 23, "condition1_3" -> 30,
"condition2_1" -> 28,
"condition2_2" -> 23|>, <|"Genes" -> "ENSMUSG00000102693",
"condition1_1" -> 2, "condition1_2" -> 3, "condition1_3" -> 0,
"condition2_1" -> 2, "condition2_2" -> 3|>}






