I am trying to use FindClusters to segment data points into similar numbers but so far I couldn't get it work for this example:
l = {110, 111, 115, 117, 251, 254, 254, 259, 399, 400, 401,
402, 542, 546, 549, 554, 660, 660, 660, 660};
FindClusters[l]
(*
-> {{110, 111, 115, 117, 251, 254, 254, 259, 399, 400, 401, 402, 542,
546, 549, 554, 660, 660, 660, 660}}
*)
If I set the N parameter (to specify: Exactly N clusters), it works:
FindClusters[l, 5]
(*
-> {{110, 111, 115, 117}, {251, 254, 254, 259},
{399, 400, 401, 402}, {542, 546, 549, 554}, {660, 660, 660, 660}}
*)
However, my intent was to use FindClusters to figure out N.

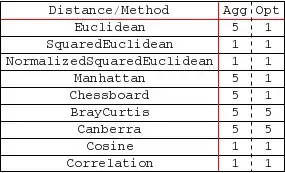
DistanceFunctionsettings?DistanceFunction -> BrayCurtisDistanceandDistanceFunction -> CanberraDistancework here, for instance... – J. M.'s missing motivation Oct 27 '12 at 17:10FindClusters[]. Mind to share more details? – Dr. belisarius Oct 27 '12 at 17:51ComponentMeasurements[ws, "BoundingBox"]
– Sven K Oct 27 '12 at 18:04mc = MorphologicalComponents@ColorNegate@i; p = Position[mc, #] & /@ Range@Max@mc; ColorNegate /@ Image /@ (SparseArray[# -> 1 & /@ Transpose[# - Min@# + 1 & /@ Transpose@#]] & /@ p)– Dr. belisarius Oct 27 '12 at 20:44