I think this is a fine question, and graphs are a great way of representing lattices, so it's natural to want to visualize using them.
UPDATE: I originally read this as a visualization question. I'll address construction at the bottom.
Visualization
Straight Render
Here's a toy ladder graph.
someGraph=Graph[{node[16], node[15], node[14], node[4], node[12], node[13], node[11], node[3], node[9], node[6], node[10], node[2], node[8], node[7],
node[1], node[5]}, {UndirectedEdge[node[16], node[15]], UndirectedEdge[node[14], node[4]], UndirectedEdge[node[16], node[14]],
UndirectedEdge[node[12], node[13]], UndirectedEdge[node[11], node[12]], UndirectedEdge[node[3], node[11]],
UndirectedEdge[node[13], node[15]], UndirectedEdge[node[12], node[16]], UndirectedEdge[node[11], node[14]],
UndirectedEdge[node[3], node[4]], UndirectedEdge[node[15], node[9]], UndirectedEdge[node[6], node[13]],
UndirectedEdge[node[10], node[2]], UndirectedEdge[node[8], node[9]], UndirectedEdge[node[10], node[8]], UndirectedEdge[node[7], node[1]],
UndirectedEdge[node[5], node[7]], UndirectedEdge[node[6], node[5]], UndirectedEdge[node[1], node[2]], UndirectedEdge[node[7], node[10]],
UndirectedEdge[node[5], node[8]], UndirectedEdge[node[6], node[9]]}]
It looks like this:

You care about correlations between adjacent (connected) nodes, I'll assume scaled between -1 and 1, and I'll generate some data so there'll be some strongly and weakly weighted edges:
(correlationWeight[#] =
RandomChoice[{-1, 1}]*(-RandomInteger[{0, 2}] 80/110 + 1);
correlationWeight[# /.
UndirectedEdge[a_, b_] :> UndirectedEdge[b, a]] =
correlationWeight[#]) & /@ EdgeList[someGraph];
Note I make sure correlationWeight is defined no matter which order someone hands me an UndirectedEdge.
We'll need an edge rendering function to care about the thickness, and color:
efStraight[pts_List,
edge_] := {Thickness[Abs[correlationWeight[edge]]/40],
If[Sign[correlationWeight[edge]] > 0, Black, Blue], Line[pts]}
and
Graph[someGraph,
VertexShapeFunction -> ({EdgeForm[{Thickness[.04], Black}],
Disk[#, .02],
EdgeForm[{Thickness[.015], White}], Disk[#, .02]} &),
EdgeShapeFunction -> efStraight]
produces:

Hopefully this gives you a good idea of the salient features.
Streaky Render
For fun I thought I'd play around for a few seconds trying to reproduce your hand-drawn effect.
A natural first attempt is to just run it through Simon Wood's ever popular:
xkcdDistort[p_] :=
Module[{r, ix, iy},
r = ImagePad[Rasterize@p, 10, Padding -> White];
{ix, iy} =
Table[RandomImage[{-1, 1}, ImageDimensions@r]~ImageConvolve~
GaussianMatrix[10], {2}];
ImagePad[
ImageTransformation[
r, # + 15 {ImageValue[ix, #], ImageValue[iy, #]} &,
DataRange -> Full], -5]];
giving us:

That's great, but everything's all rasterized, and I was curious about reproducing the streaks from the repeated marker strokes. Here's a first attempt borrowing the BSpline wiggles from Mr. Wizard :
efStreaky[pts_List, e_] := {Thickness[Abs[correlationWeight[e]]/40],
If[Sign[correlationWeight[e]] > 0, Opacity[.4, Black],
Opacity[.5, RGBColor[0., 0.26, 0.79]]],
setBackLineJiggle[pts, xx = RandomReal[{1/8, 1/5}]],
setBackLineJiggle[pts, xx*1.1 ]}
Graph[someGraph,
VertexShapeFunction -> ({EdgeForm[{Thickness[.04], Black}],
Disk[#, .02],
EdgeForm[{Thickness[.015], White}], Disk[#, .02]} &),
EdgeShapeFunction -> efStreaky]
yielding

and

with wiggle helper code:
split[{a_, b_}] :=
If[a == b, {b},
With[{n = Ceiling[3 Norm[a - b]]},
Array[{n - #, #}/n &, n].{a, b}]];
partition[{x_, y__}] := Partition[{x, x, y}, 2, 1];
nudge[L : {a_, b_}, d_] := Mean@L + d Cross[a - b];
wiggle[pts : {{_, _} ..},
d_: {-0.15, 0.15}] := ## &[#~nudge~RandomReal@d, #[[2]]] & /@
partition[Join @@ split /@ partition@pts];
setBackLineJiggle[{a_, b_}, n_] := BSplineCurve@wiggle@{a + n (b - a) +
RandomReal[{-1, 1}, 2] /(30 Norm[b - a]), b - n (b - a)}
Graph Construction
From Correlation Data
Somehow you have to be starting with some correlation data between lattice sites. One way you could be storing it is a sparse array that has values when sites have non-vanishing correlation between them.
Something like a sparse version of:
correlationTable = {{0, -1, 0, 0, 0, 0, -(3/11), 0, 0, 0, 0, 0, 0, 0,
0, 0}, {-1, 0, 0, 0, 0, 0, 0, 0, 0, -(3/11), 0, 0, 0, 0, 0, 0}, {0,
0, 0, 5/11, 0, 0, 0, 0, 0, 0, -1, 0, 0, 0, 0, 0}, {0, 0, 5/11, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 3/11, 0, 0}, {0, 0, 0, 0, 0, 3/11, 1, 5/
11, 0, 0, 0, 0, 0, 0, 0, 0}, {0, 0, 0, 0, 3/11, 0, 0, 0, 1, 0, 0,
0, -1, 0, 0, 0}, {-(3/11), 0, 0, 0, 1, 0, 0, 0, 0, -1, 0, 0, 0, 0,
0, 0}, {0, 0, 0, 0, 5/11, 0, 0, 0, -(3/11), 5/11, 0, 0, 0, 0, 0,
0}, {0, 0, 0, 0, 0, 1, 0, -(3/11), 0, 0, 0, 0, 0, 0, -(3/11),
0}, {0, -(3/11), 0, 0, 0, 0, -1, 5/11, 0, 0, 0, 0, 0, 0, 0, 0}, {0,
0, -1, 0, 0, 0, 0, 0, 0, 0, 0, -(5/11), 0, -(3/11), 0, 0}, {0, 0,
0, 0, 0, 0, 0, 0, 0, 0, -(5/11), 0, 1, 0, 0, 3/11}, {0, 0, 0, 0,
0, -1, 0, 0, 0, 0, 0, 1, 0, 0, 3/11, 0}, {0, 0, 0, 3/11, 0, 0, 0,
0, 0, 0, -(3/11), 0, 0, 0, 0, 1}, {0, 0, 0, 0, 0, 0, 0, 0, -(3/11),
0, 0, 0, 3/11, 0, 0, 1}, {0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 3/11,
0, 1, 1, 0}};
i.e.
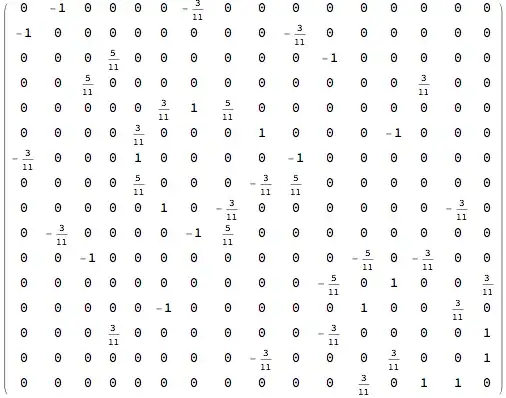
It's very easy to turn this data, or it's sparse version, into a graph, connecting only edges with non-vanishing correlation.
Here's one way, that builds the correlationWeight function used above in one go:
someGraph2 = (SparseArray[correlationTable] //
ArrayRules)[[1 ;; -2]] /.
Rule[{a_, b_},
weight_] :> (correlationWeight[
UndirectedEdge[node[a], node[b]]] = weight;
correlationWeight[UndirectedEdge[node[b], node[a]]] = weight;
UndirectedEdge @@ Sort[{node[a], node[b]}]) // Union // Graph







GridGraphobjects. It would be valuable to have some insight into an intelligent way of doing this. – Diffycue Aug 14 '17 at 23:59Graphfor this? If the structure is always built from simple repeating ladders like yours, why don't you write a visualization yourself? When I see this right, you only need points and bonds of different thickness. Everything else can be built upon these basic structures. – halirutan Aug 15 '17 at 01:25