So far i have managed to implement the Crude Monte Carlo (CMC) and the Importance Sample Monte Carlo (ISMC) for unidimensional problems.
Consider the function func[x_] := Exp[-2 Abs[x - 5]]
The CMC formulation is given by:
$I[f]=E_{p}[f(x)]=\int{f(x)p(x)}dx \approx \frac{1}{N} \sum_{i=1}^{N}{f(x^{i})} $
The code below perform the integration NIntegrate[func[x], {x, 0, 10}]
func[x_] := Exp[-2 Abs[x - 5]]
nsamples = 10000;
X = RandomVariate[UniformDistribution[{0, 10}], nsamples];
Y[X_] := 10 func[X]
vals = Table[Y[X[[i]]], {i, 1, Length[X]}];
Print["Integration crude Monte Carlo = ", Mean[vals]]
Print["Sample variance crude Monte Carlo = ", Variance[vals]]
Integration crude Monte Carlo = 1.00616
Sample variance crude Monte Carlo = 4.04767
$I[f]=E_{p}[f(x)]=\int{f(x)\frac{p(x)}{q(x)} q(x)}dx \approx \frac{1}{N} \sum_{i=1}^{N}{\frac{p(x^{i})}{q(x^{i})} f(x^{i})} $
$\hat{I}[f]=\sum_{i=1}^{N}{ w f(x^{i})} , w=\frac{p(x^{i})}{q(x^{i})} $
and ISMC solution:
W[X_] := PDF[UniformDistribution[{0, 10}], X]/
PDF[NormalDistribution[5, 1], X]
f[X_] := 10 func[X]
X = RandomVariate[NormalDistribution[5, 1], nsamples];
vals = Table[W[X[[i]]] f[X[[i]]], {i, 1, Length[X]}];
Print["Integration Importance Sample Monte Carlo = ", Mean[vals]]
Print["Sample variance Importance Sample Monte Carlo = ",
Variance[vals]]
Integration Importance Sample Monte Carlo = 0.998383
Sample variance Importance Sample Monte Carlo = 0.356451
Note that the problem is 1D. Now I need to implement the ISMC for 2D problems.
2D Example:
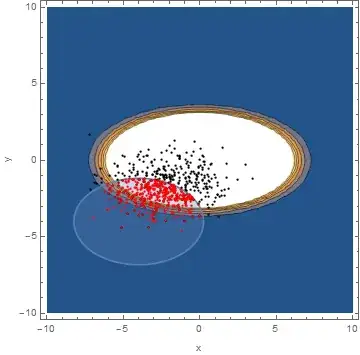
In order to obtain a failure probability of an event, I need to integrate the PDF[BinormalDistribution[{0, 0}, {2, 1}, 0], {x, y}] over a region. In my case this region is defined by the following limit state function:
gx[x_, y_] := (((y + 4)/2)^2 + ((x + 4)/3)^2 - 2)
I can compute compute the exact failure probability of this event and also it's reliabily index using NIntegrate:
mean0 = 0.;
mean1 = 0.;
sdev0 = 2;
sdev1 = 1;
cov = 0;
gx[x_, y_] := (((y + 4)/2)^2 + ((x + 4)/3)^2 - 2)
cp = ContourPlot[{PDF[BinormalDistribution[{mean0, mean1}, {sdev0, sdev1},cov], {x, y}]}, {x, -10, 10}, {y, -10, 10}, PerformanceGoal ->"Quality",Contours -> 10];
rp = RegionPlot[gx[x, y] < 0, {x, -10, 10}, {y, -10, 10}];
reg = ImplicitRegion[
gx[x, y] < 0, {{x, -Infinity, Infinity}, {y, -Infinity, Infinity}}];
Show[cp, rp]
exactpf = NIntegrate[PDF[BinormalDistribution[{mean0, mean1}, {sdev0, sdev1}, cov], {x, y}], Element[{x, y}, reg]];
reliabilityindex = -InverseCDF[NormalDistribution[0, 1], exactpf];
Print["Exact probability = ", exactpf]; Print["Exact reliability index = ", reliabilityindex];
`Exact probability = 0.0234098
Exact reliability index = 1.98793`
How can I solve the above 2D problem using ISMC?