It might make sense to use a few sinusoids instead of e.g. Gaussians. While there are very likely better ways to go about this using windowing, I show a naive approach where we simply clip frequencies that do not have large amplitudes.
data = {{-4053, 0.0970776}, {-4052.9, 0.105458}, {-4052.8,
0.120125}, {-4052.7, 0.136886}, {-4052.6, 0.14841}, {-4052.5,
0.14806}, {-4052.4, 0.123966}, {-4052.3, 0.107903}, {-4052.2,
0.0869506}, {-4052.1, 0.0625067}, {-4052, 0.0523801}, {-4051.9,
0.042253}, {-4051.8, 0.0359675}, {-4051.7, 0.0314279}, {-4051.6,
0.0293327}, {-4051.5, 0.0296819}, {-4051.4, 0.0289835}, {-4051.3,
0.0324755}, {-4051.2, 0.0338723}, {-4051.1, 0.0426022}, {-4051,
0.049237}, {-4050.9, 0.0635543}, {-4050.8, 0.0841568}, {-4050.7,
0.0984741}, {-4050.6, 0.118728}, {-4050.5, 0.127457}, {-4050.4,
0.133743}, {-4050.3, 0.1306}, {-4050.2, 0.0981248}, {-4050.1,
0.0893951}, {-4050, 0.0747286}, {-4049.9, 0.0555226}, {-4049.8,
0.0464437}, {-4049.7, 0.0384118}, {-4049.6, 0.0321263}, {-4049.5,
0.0310787}, {-4049.4, 0.0293327}, {-4049.3, 0.0293327}, {-4049.2,
0.0293327}, {-4049.1, 0.0289835}, {-4049, 0.0415546}, {-4048.9,
0.0408562}, {-4048.8, 0.0495863}, {-4048.7, 0.0740302}, {-4048.6,
0.0813634}, {-4048.5, 0.0963792}, {-4048.4, 0.120823}, {-4048.3,
0.13514}, {-4048.2, 0.140029}, {-4048.1, 0.127807}, {-4048,
0.12222}, {-4047.9, 0.103712}, {-4047.8, 0.0796173}, {-4047.7,
0.0677446}, {-4047.6, 0.0593636}, {-4047.5, 0.0478401}, {-4047.4,
0.0419038}, {-4047.3, 0.0366659}, {-4047.2, 0.0331739}, {-4047.1,
0.0310787}, {-4047, 0.0335231}, {-4046.9, 0.0408562}, {-4046.8,
0.0433006}, {-4046.7, 0.0457451}, {-4046.6, 0.0625067}, {-4046.5,
0.068443}, {-4046.4, 0.0820619}, {-4046.3, 0.099871}, {-4046.2,
0.119077}, {-4046.1, 0.13514}, {-4046, 0.131997}, {-4045.9,
0.132695}, {-4045.8, 0.118029}, {-4045.7, 0.0859029}, {-4045.6,
0.0740302}, {-4045.5, 0.0604113}, {-4045.4, 0.0516816}, {-4045.3,
0.0394594}, {-4045.2, 0.0342215}, {-4045.1, 0.0321263}, {-4045,
0.0307295}, {-4044.9, 0.0303803}, {-4044.8, 0.0293327}, {-4044.7,
0.0338723}, {-4044.6, 0.0384118}, {-4044.5, 0.0412054}, {-4044.4,
0.0534273}, {-4044.3, 0.0698399}, {-4044.2, 0.0810142}, {-4044.1,
0.109998}, {-4044, 0.126061}, {-4043.9, 0.137934}, {-4043.8,
0.133394}, {-4043.7, 0.133743}, {-4043.6, 0.120125}, {-4043.5,
0.0900936}, {-4043.4, 0.084506}, {-4043.3, 0.0691415}, {-4043.2,
0.0548242}, {-4043.1, 0.0506339}, {-4043, 0.0429514}, {-4042.9,
0.0391102}, {-4042.8, 0.0384118}, {-4042.7, 0.0380627}, {-4042.6,
0.0426022}, {-4042.5, 0.0457451}, {-4042.4, 0.0488878}, {-4042.3,
0.0663477}, {-4042.2, 0.0673953}, {-4042.1, 0.0771727}, {-4042,
0.113839}, {-4041.9, 0.126759}, {-4041.8, 0.144568}, {-4041.7,
0.158536}, {-4041.6, 0.159235}, {-4041.5, 0.153298}, {-4041.4,
0.13095}, {-4041.3, 0.108252}, {-4041.2, 0.0824106}, {-4041.1,
0.0653}, {-4041, 0.0548242}, {-4040.9, 0.0471421}, {-4040.8,
0.0394594}, {-4040.7, 0.0363167}, {-4040.6, 0.0335231}, {-4040.5,
0.0359675}, {-4040.4, 0.0359675}, {-4040.3, 0.0412054}, {-4040.2,
0.0457451}, {-4040.1, 0.0534273}, {-4040, 0.0663477}, {-4039.9,
0.0872998}, {-4039.8, 0.103712}, {-4039.7, 0.12641}, {-4039.6,
0.156092}, {-4039.5, 0.17006}, {-4039.4, 0.16971}, {-4039.3,
0.159933}, {-4039.2, 0.124664}, {-4039.1, 0.10476}, {-4039,
0.0869506}, {-4038.9, 0.0670461}, {-4038.8, 0.0579672}, {-4038.7,
0.0506339}, {-4038.6, 0.0446976}, {-4038.5, 0.0415546}, {-4038.4,
0.0429514}, {-4038.3, 0.0443482}, {-4038.2, 0.0443482}, {-4038.1,
0.0506339}, {-4038, 0.0635543}, {-4037.9, 0.0691415}, {-4037.8,
0.084506}, {-4037.7, 0.114887}, {-4037.6, 0.128854}, {-4037.5,
0.149806}, {-4037.4, 0.166568}, {-4037.3, 0.176345}, {-4037.2,
0.170409}, {-4037.1, 0.133394}, {-4037, 0.11768}, {-4036.9,
0.0981248}, {-4036.8, 0.0733317}, {-4036.7, 0.0579672}, {-4036.6,
0.0520308}, {-4036.5, 0.043999}, {-4036.4, 0.0412054}, {-4036.3,
0.0391102}, {-4036.2, 0.0342215}, {-4036.1, 0.0387611}, {-4036,
0.0398087}, {-4035.9, 0.0509832}, {-4035.8, 0.0516816}, {-4035.7,
0.0632051}, {-4035.6, 0.0949823}, {-4035.5, 0.108601}, {-4035.4,
0.129902}, {-4035.3, 0.154695}, {-4035.2, 0.172504}, {-4035.1,
0.177742}, {-4035, 0.158536}, {-4034.9, 0.142473}, {-4034.8,
0.115934}, {-4034.7, 0.0820619}, {-4034.6, 0.068443}, {-4034.5,
0.0555226}, {-4034.4, 0.0457451}, {-4034.3, 0.0391102}, {-4034.2,
0.0377134}, {-4034.1, 0.0352691}, {-4034, 0.0363167}, {-4033.9,
0.0356183}, {-4033.8, 0.0415546}, {-4033.7, 0.043999}, {-4033.6,
0.0530785}, {-4033.5, 0.0642528}, {-4033.4, 0.0960299}, {-4033.3,
0.109648}, {-4033.2, 0.128156}, {-4033.1, 0.138981}, {-4033,
0.152251}, {-4032.9, 0.151901}, {-4032.8, 0.128505}, {-4032.7,
0.10441}, {-4032.6, 0.0799665}, {-4032.5, 0.0604113}, {-4032.4,
0.0467929}, {-4032.3, 0.0384118}, {-4032.2, 0.0279359}, {-4032.1,
0.0233964}, {-4032, 0.0261899}};
ft = Fourier[data[[All, 2]]];
Lets see what the spectrum looks like in terms of magnitudes.
ListPlot[Abs[ft]]

We'll clip at magnitude 0.05.
clipped = ft /. (aa_ /; Abs[aa] <= .05 :> 0);
ListPlot[Abs[clipped]]

Now take the inverse FT of the clipped FT to get the low dimensional (in terms of number of frequencies) approximation.
approx = Re[InverseFourier[clipped]];
We superimpose list plots to check by eye that this gave a reasonable approximation.
ListPlot[{approx, data[[All, 2]]}]





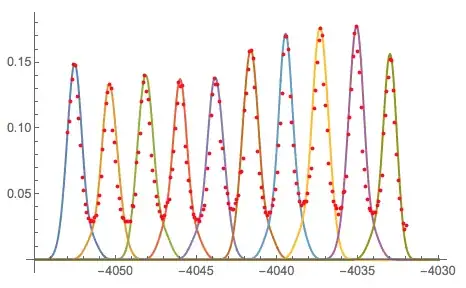





cutSlowAxisis not defined in the question. – Anton Antonov Jun 25 '18 at 02:10Fourierand zero all but the largest components, the result ofInverseFourieris still a fairly good approximation. – Daniel Lichtblau Jun 28 '18 at 23:11