Partial ND
It's possible to again extract all this weight data and stuff in the higher dimensional cases using effectively the same procedure as for the 1D:
fdmWeightDataND~SetAttributes~HoldAll;
fdmWeightDataND[a_, dep_Symbol, indeps : {__Symbol}, hs : {__Symbol}] :=
Block[{dep}, Block[indeps, Block[hs,
Module[{weights, order, denomWeight, denom, num, powed, pows, minpow,
pts, x}, denom = Denominator[a];
(*assumes the mesh spacing is in there at different powers*)
order = Exponent[denom, #] & /@ hs;
(*pull the power on the mesh spacing*)
denomWeight = If[order > 0, Coefficient[denom, hs^order], denom];
num = Numerator[a];
{powed, pows} =
Reap[
num /.
dep[args___]?(Not@*FreeQ[Alternatives @@ indeps]) :>
dep @@
Sow[
MapThread[
Coefficient[#, #2] &,
{
{args},
hs
}
]
]
];
minpow = Min[pows];
weights =
AssociationThread[
pows[[1]],
Map[Coefficient[powed /. (dep @@ #) -> x, x] &, pows[[1]]]
];
<|
"Weights" -> weights,
"Orders" -> order
|>
]
]
]
];
This is less automate-able, though, since you need to specify what your f, independent variables, and mesh spacings are. Also without knowing how you'd like to flatten your nD tensor of grid points it's impossible to automatically build the FD matrix. But at a minimum I can give you weights and indices for the weights:
(*didn't want to Wikipedia the nine-point stencil so most of this is fake*)
simpleFormula = (
f[x - xh, y] + f[x + xh, y] +
f[x, y - yh] + f[x, y + yh] - 4 f[x, y] +
1/2 f[x + xh, y - yh] + 1/2 f[x - xh, y - yh] +
1/2 f[x + xh, y + yh] + 1/2 f[x - xh, y + yh]
)/(xh^2*yh^2);
weights2D = fdmWeightDataND[simpleFormula, f, {x, y}, {xh, yh}]
<|"Weights" -> <|{0, 0} -> -4, {0, -1} -> 1, {0, 1} -> 1, {-1, 0} ->
1, {-1, -1} -> 1/2, {-1, 1} -> 1/2, {1, 0} -> 1, {1, -1} -> 1/2, {1, 1} ->
1/2|>, "Orders" -> {2, 2}|>
Basically now you'd pick how to order your grid points and build a matrix from that. Here's one way to do this based on having the second index be the slowest moving (note that I'm only like 50% confident I built this correctly):
FDM2D[weightData_, {hx_, hy_}, {nptsx_, nptsy_}] :=
Module[{xps, yps, weights = weightData["Weights"],
orders = weightData["Orders"]},
xps = Length@DeleteDuplicates@Keys[weights][[All, 1]];
yps = Length@Sort@DeleteDuplicates@ Keys[weights][[All, 2]];
SparseArray[
Flatten@
KeyValueMap[
With[{ix = #[[1]], iy = #[[2]], v = #2},
Map[
Band[
If[ix >= 0, {1, 1 + ix}, {1 - ix, 1}] + {#, #} +
If[iy >= 0, {0, iy*nptsx}, {-iy*nptsx, 0}],
{nptsx*nptsy, nptsx*nptsx},
{nptsx, nptsx}
] -> v &,
Range[nptsx - Abs[ix]] - 1
]
] &,
weights
],
{nptsx*nptsy, nptsx*nptsx}
]/(Times @@ ({hx, hy}^orders))
]
Here's what that outputs:
FDM2D[weights2D, {1, 1}, {5, 5}] // MatrixPlot
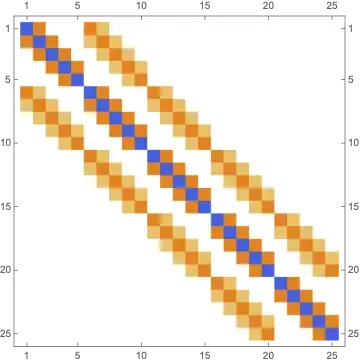
In the case of no coupling across degrees of freedom this looks correct I believe... (but can't be sure since I usually do my Laplacian via a Kronecker sum):
laplacian = (
f[x - xh, y] + f[x + xh, y] +
f[x, y - yh] + f[x, y + yh] - 4 f[x, y]
)/(xh^2*yh^2);
FDM2D[fdmWeightDataND[laplacian, f, {x, y}, {xh, yh}], {1, 1}, {10,
10}] // MatrixPlot

Full Thing for 1D
Here's a method that automates this extraction for 1D FDM stuff:
fdmWeightData~SetAttributes~HoldAll;
fdmWeightData[Equal[a_, b_], dep : {__Symbol}, indep : {__Symbol}] :=
fdmWeightData[a, dep, indep];
fdmWeightData[a_, dep_Symbol, indep_Symbol, h_Symbol] :=
Block[{dep, indep, h},
Module[{weights, order,
denomWeight, denom,
num, powed, pows,
minpow, pts
},
denom = Denominator[a];
(* assumes the mesh spacing is in there *)
order = Exponent[denom, h];
(* pull the power on the mesh spacing *)
denomWeight = Coefficient[ denom, h^order];
num = Numerator[a];
{powed, pows} =
Reap[
num /.
dep[expr_?(Not@*FreeQ[indep])] :>
dep[Sow@Coefficient[expr, h]]
];
minpow = Min[pows];
powed = powed /. dep[n_] :> dep^(n - minpow);
weights = (CoefficientList[powed, dep])/denomWeight;
pts = Exponent[powed, dep, List] + minpow;
<|
"Weights" -> weights,
"Points" -> pts,
"Order" -> order
|>
]
];
fdmWeightData[Equal[expr_, b_]] :=
fdmWeightData[expr];
fdmWeightData[expr_] :=
Module[
{
vars = Variables[expr],
dep,
indep,
h
},
h = FirstCase[vars, _Symbol];
{dep, indep} =
Replace[
FirstCase[vars, a_Symbol[b_Symbol] :> {a, b}, None],
None :>
a_Symbol[e_] :> FirstCase[e, Except[h, _Symbol], None, Infinity]
] ;
With[
{dep = dep, indep = indep, h = h},
fdmWeightData[expr, dep, indep, h]
]
];
fdmWeightData[(y[x + dx] - 2 y[x] + y[x - dx])/dx^2 == Sin[x]]
<|"Weights" -> {1, -2, 1}, "Points" -> {-1, 0, 1}, "Order" -> 2|>
Here's a helper on that:
fdmMat[weights_List, pts_List, order_List, h_, n_] :=
SparseArray[
MapThread[
If[# >= 0,
Band[{1, # + 1}] -> #2,
Band[{-# + 1, 1}] -> #2
] &,
{pts, weights/(h^order)}
],
{n, n}
];
fdmMat[a_Association, h_, n_] :=
fdmMat[Sequence @@ Lookup[a, {"Weights", "Points", "Order"}], h, n];
fdmMat[a_Association _, n_] :=
fdmMat[a, 1, n];
FDMMatrix[a_Association][n_, h_: 1] :=
fdmMat[a, h, n];
FDMMatrix[expr : Except[_Pattern | _Association]] :=
FDMMatrix[fdmWeightData[expr]]
FDMMatrix[expr_, dep_, indep_, h_] :=
FDMMatrix[fdmWeightData[expr, dep, indep, h]]
fdmat = FDMMatrix[(y[x + dx] - 2 y[x] + y[x - dx])/dx^2]
FDMMatrix[<|"Weights" -> {1, -2, 1}, "Points" -> {-1, 0, 1}, "Order" -> 2|>]
Then we can make the matrix fast:
fdmat[100] // MatrixPlot

Doing the nD extension will take a bit of thought, though.
Partial Solution to Actual Problem
I'm not sure how your actual problem works, but here's a way we can try to build an FD matrix for it using this stuff. The main thing we'll do is break it down into portions with the same dx and dy terms after doing some symbolic pre-simplification. Here's my new setup (same as yours but entirely symbolic):
Clear[r, glass, air, k]
n2 = (Function[{x, y}, #] &@(Simplify`PWToUnitStep@
PiecewiseExpand@If[x^2 + y^2 <= r^2, glass, air]));
ClearAll[fw, bw, delta]
SetAttributes[#, HoldAll] & /@ {fw, bw};
fw@D[expr_, x_] :=
With[{\[CapitalDelta] = delta@ToString@x},
Subtract @@ (expr /. {{x -> x + \[CapitalDelta]}, {x -> x}})/\[CapitalDelta]]
bw@D[expr_, x_] :=
With[{\[CapitalDelta] = delta@ToString@x},
Subtract @@ (expr /. {{x -> x}, {x -> x - \[CapitalDelta]}})/\[CapitalDelta]]
delta[a_ + b_] := delta@a + delta@b
delta[k_. delta[_]] := 0
grad = Function[expr, fw@D[expr, #] & /@ {x, y}];
div = Function[expr, Total@MapThread[bw@D@## &, {expr, {x, y}}]];
elst = e[#][x, y] & /@ Range[2];
baseDiscreteLHS =
With[{n2 = n2[x, y]},
div@grad@elst + (n2 k^2) elst - grad[div@elst - 1/n2 div[n2 elst]]];
Now I'll prep this stuff so it's in a form my code can use cleanly:
prepped =
(baseDiscreteLHS[[1]]) //. {
e[1] -> e1,
e[2] -> e2,
delta["y"] -> dy,
delta["x"] -> dx,
UnitStep[r^2 - x^2 - y^2] -> usxy,
UnitStep[r^2 - x^2 - (-dy + y)^2] -> usxdy,
UnitStep[r^2 - (dx + x)^2 - y^2] -> usdxy,
UnitStep[r^2 - (dx + x)^2 - (-dy + y)^2] -> usdxdy,
UnitStep[r^2 - (-dx + x)^2 - y^2] -> usdxmdy
} // FullSimplify;
Now I'll use this stuff to pull the FD weights just for e1 from the first of the two equations provided:
weightsE1 =
{
fdmWeightDataND[prepped[[1]], e1, {x, y}, {dx, dy}],
fdmWeightDataND[prepped[[2]], e1, {x, y}, {dx, dy}],
fdmWeightDataND[prepped[[3]], e1, {x, y}, {dx, dy}],
fdmWeightDataND[prepped[[4]], e1, {x, y}, {dx, dy}],
fdmWeightDataND[
Simplify[Collect[prepped[[6]] // Expand, dx][[1]]],
e1,
{x, y},
{dx, dy}
],
fdmWeightDataND[
Simplify[Collect[prepped[[8]] // Expand, dx][[1]]],
e1,
{x, y},
{dx, dy}
]
};
Then I'll build all the matrices, add them together, and numericize everything:
mate1 =
Block[
{
r = .8, glass = (1.45)^2, air = 1., k = (2 \[Pi])/1.55,
sp1, spRules, points = 25,
dx, dy
},
dx = 1./points; dy = 1/points;
sp1 = FDM2D[#, {dx, dy}, {points, points}] & /@ weightsE1 // Total;
spRules =
MapThread[
# ->
With[{x = #[[1]]*dx, y = #[[2]]*dy},
#2 /.
{
usxy -> UnitStep[r^2 - x^2 - y^2],
usxdy -> UnitStep[r^2 - x^2 - (-dy + y)^2],
usdxy -> UnitStep[r^2 - (dx + x)^2 - y^2],
usdxdy -> UnitStep[r^2 - (dx + x)^2 - (-dy + y)^2],
usdxmdy -> UnitStep[r^2 - (-dx + x)^2 - y^2]
}
] &,
{
sp1["NonzeroPositions"],
sp1["NonzeroValues"]
}
];
SparseArray[spRules, Dimensions[sp1]]
]
Finally we can solve this using your same Eigensystem code:
Block[{r = .8, glass = (1.45)^2, air = 1., k = (2 \[Pi])/1.55, points = 25},
{val, vec} =
Eigensystem[mate1, -6, Method -> {"Arnoldi", "Shift" -> k^2 glass}
];
mat = ArrayReshape[#, {2, points - 2, points - 2}] & /@ vec;
MapThread[
ArrayPlot[#[[1]], PlotLabel -> Sqrt@#2, PlotRange -> All,
ColorFunction -> "AvocadoColors"] &,
{mat, val}
]
] // AbsoluteTiming
The results are meaningless since I just randomly took part of the problem, but this entire procedure was fast and not memory intensive at all.







e[1]ande[2], so for which of these are we trying to get the difference matrix... I am not much of a DEs aficionado so I'm not sure what the "standard" way to treat that coupling in the FD formulation is. – b3m2a1 Jun 27 '19 at 19:41vare. – xzczd Jun 28 '19 at 02:21n2 = Function[{x, y}, If[x^2 + y^2 <= r^2, glass, air]], then time is reduced by 2 times ;) – Alex Trounev Jun 28 '19 at 11:38UnitStepprobably won't help (because there's no vectorization), but never expected it actually slows down the code in this case! Code improved. Thanks for pointing out! – xzczd Jun 28 '19 at 12:15//N(this is not necessary, since the parameters are not exactly defined) and replaceBlockwithModule. – Alex Trounev Jun 28 '19 at 17:52//Nimproves the performance. Actually I add it to handlegridbecause in certain cases complicated symbolic grid can slow down the code, but I didn't expect inexact grid is more complicated in this case. Edited, thx for pointing out. However,Blockshould not be replaced byModule, or the Dirichlet b.c. won't be set. Just tryexpr = f[4]; Module[{f}, f[x_] =x^2; expr]. The code still produces reasonable result because b.c. isn't that important for this problem. Another example is here: https://mathematica.stackexchange.com/q/163486/1871 – xzczd Jun 29 '19 at 02:52e[i_][L | -L, _] = e[i_][_, L | -L] = 0.;is just unnecessary, because the zero Dirichlet b.c. has already been set bydel@gridinvare. (Since the elements at the boundary isn't included invare,CoefficientArraywill treat these elements as constants and move them tobmrray2. This is equivalent to setting zero Dirichlet b.c.. ) – xzczd Jun 24 '20 at 04:19