Method #1: Image Processing
Find intersections
PerpBis[pt1_, pt2_] :=
Block[{mid = Midpoint[{pt1, pt2}], vec = pt2 - pt1, gamma},
gamma = -vec[[1]]*mid[[1]] - vec[[2]]*mid[[2]];
vec[[1]]*x + vec[[2]]*y + gamma == 0]
crossings =
Table[{x, y} /.
Solve[PerpBis[{0, 0}, {x1, y1}] &&
PerpBis[{0, 0}, {x2, y2}]], {x1, -2, 2}, {y1, -2, 2}, {x2, -2,
2}, {y2, -2, 2}];
crossingsFiltered =
Cases[Flatten[crossings, 4], {_?NumericQ, _?NumericQ}] //
DeleteDuplicates;
Create two plots
We add line intersections as big black points to the first plot img which will help us determine the neighbouring regions. The second one will be used to produce the final image. Both plots should be created equally so that they concide. ImageSize should be large enough so that even the smallest zones are visible.
out = Show[
Table[ContourPlot[
Evaluate@PerpBis[{0, 0}, {i, j}], {x, -5, 5}, {y, -5, 5},
ContourStyle -> Black], {i, -2, 2}, {j, -2, 2}],
ListPlot[crossingsFiltered, PlotStyle -> Black,
PlotMarkers -> {Automatic, 12}]];
img = Binarize@Rasterize[out, ImageSize -> 800];
plot = Rasterize[
Show@Table[
ContourPlot[
Evaluate@PerpBis[{0, 0}, {i, j}], {x, -5, 5}, {y, -5, 5},
ContourStyle -> Black], {i, -2, 2}, {j, -2, 2}],
ImageSize -> 800];
{img, plot} // GraphicsRow

Find Brillouin zones
We use MorphologicalComponents to find the zones. Then Dilation is used so that the lines disappear.
dilationSize = 10;
m = DeleteBorderComponents[
Dilation[
DeleteSmallComponents[
MorphologicalComponents[img, CornerNeighbors -> False], dilationSize], .5],
CornerNeighbors -> False];
plotComp =
DeleteBorderComponents@
DeleteSmallComponents[
MorphologicalComponents[Binarize@plot, CornerNeighbors -> False],
50];
m // Colorize
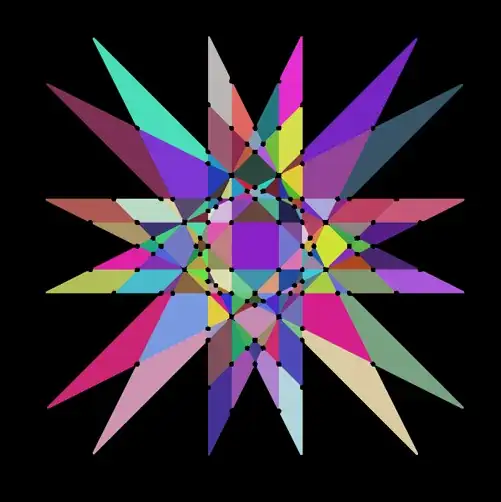
Create graph
First, we extract morphological parameters (labels, centroids and neighbors). Because we've added the black points, the neighboring Brillouin zones will be correctly identified.
nodes = ComponentMeasurements[m, "Label", CornerNeighbors -> False][[All, 1]];
centroids = ComponentMeasurements[m, "Centroid", CornerNeighbors -> False];
neighbors = ComponentMeasurements[m, "Neighbors", CornerNeighbors -> False];
g = Graph[nodes,
DeleteDuplicates[
Sort /@ Flatten[
neighbors /. (i_ -> {l___}) :> (i [UndirectedEdge] # & /@
{l})]], VertexCoordinates -> centroids, VertexSize -> .5,
VertexStyle -> Red];
Show[img, g]
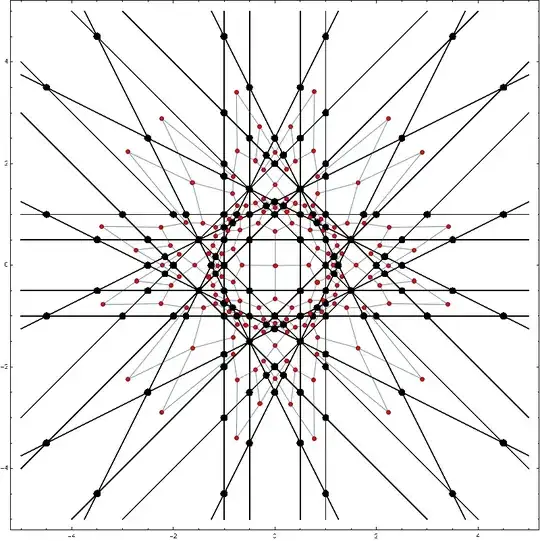
Calculate distances
The central Brillouin zone corresponds to the center of graph (smallest eccentricity). Then we find a mapping between morphological components of both plots.
centerNode = First@GraphCenter[g];
distance = GraphDistance[g, centerNode];
centroidsRounded = #[[1]] -> Round@#[[2]] & /@ centroids;
plotRules =
MapThread[
plotComp[[-#1[[2, 2]], #1[[2, 1]]]] -> #2 + 1 &, {centroidsRounded,
distance}];
Create plot
First we hide the areas around the Brillouin zones. Then we use some fancy color scheme.
finalPlot = plotComp /. {0 -> -1, 1 -> -1};
finalPlot =
ColorReplace[
Colorize[Replace[finalPlot, plotRules, 2],
ColorFunction -> ColorData[93], ColorFunctionScaling -> False],
Black -> Transparent]
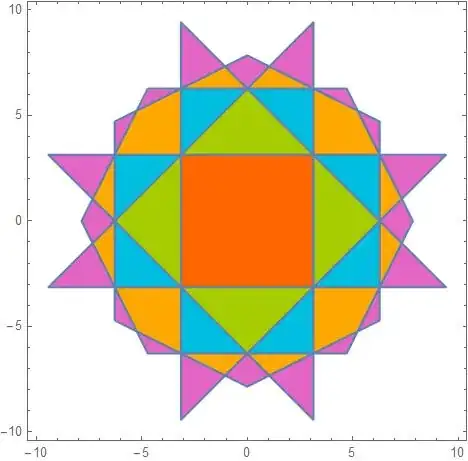
ImageCompose[plot, finalPlot]

Warning
Because we use image processing, we have to be careful to examine the final results. If the initial image resolution is too small, some zones might get missed. If the intersections are not plotted large enough or dilationSize is too large, some zones may get wrongfully connected as neighbours.
Method #2: Dual Graph
Why bother with image processing when we can calculate the exact positions of all the vertices and edges.
Generate graph
lines = (Table[PerpBis[{0, 0}, {x1, y1}], {x1, -2, 2}, {y1, -2, 2}] //
FullSimplify // Flatten) /. True -> Nothing;
crossings = ({x, y} /. Solve[#[[1]] && #[[2]]]) & /@
Tuples[lines, {2}];
nodes = Cases[Flatten[crossings, 1], {_?NumericQ, _?NumericQ}] //
DeleteDuplicates;
(* Connect neighbouring points on the same line *)
generateEdges[pts_] := Partition[SortBy[pts, nodes[[#]] &], 2, 1];
(* Generate all possible edges *)
edgesAll =
Flatten[generateEdges[
Pick[Range@Length@nodes,
Function[p, # /. {x -> p[[1]], y -> p[[2]]}] /@ nodes]] & /@
lines, 1];
(* Remove self-edges and turn lists into undirected edges *)
edges = Select[edgesAll, #[[1]] != #[[2]] &] /. {a_, b_} :>
a [UndirectedEdge] b // DeleteDuplicates;
G0 = Graph[Range[Length@nodes], edges, VertexCoordinates -> nodes]

Find dual graph
There seems to be no build-in function to generate a dual graph. So we will generate it ourselves. First, we find faces with IGraph, but this can also be done without any packages. Then, we compare the edges and find the neighbouring faces.
<< IGraphM`
(* Find faces *)
faces0 = IGFaces[G0];
(* Remove the outer-most face *)
faces = Sort[faces0][[1 ;; -2]];
(* Calculate centroids *)
facesCenter = Mean[Part[nodes, #]] & /@ faces;
(* Make the first vertex appear again at the end of the list *)
facesVertices = Append[#, #[[1]]] & /@ faces;
(* Create lists of edges *)
facesEdges = (Sort /@ Partition[#, 2, 1]) & /@ facesVertices;
(* Find faces that share an edge *)
conn = Table[
If[i < j &&
Length[facesEdges[[i]] [Intersection] facesEdges[[j]]] == 1,
i [UndirectedEdge] j, Nothing], {i, 1, Length@facesEdges}, {j,
1, Length@facesEdges}] // Flatten // DeleteDuplicates;
G = Graph[Range@Length@faces, conn, VertexCoordinates -> facesCenter]

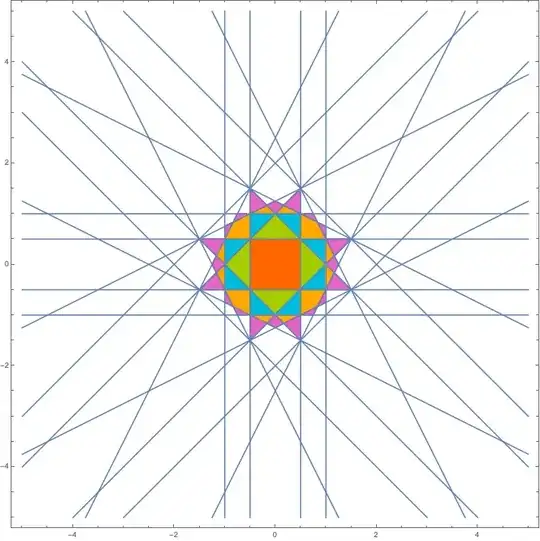
Calculate distances
This is done the same as before. However, let's now take only first few Brillouin zones.
centerNode = First@GraphCenter[G];
distances = GraphDistance[G, centerNode];
(* Maximal Brillouin zone *)
maxDistance = 7;
ind = LessThan[maxDistance] /@ distances;
brillouinZones =
Graphics[MapThread[{ColorData[106][#2],
Polygon[Part[nodes, #1]]} &, {Pick[facesVertices, ind],
Pick[distances, ind]}]];
plot = ContourPlot[Evaluate@#, {x, -5, 5}, {y, -5, 5}] & /@ lines;
Show[plot, brillouinZones]
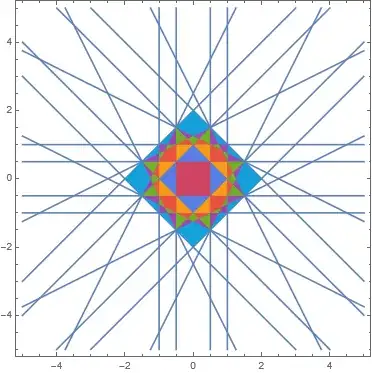
Advantages
This method has significant advantages over the image processing method because (1) it does not need any manual fine-tuning, (2) it generates polygons instead of a rasterized image, (3) it is certain to correctly find all the zones.










gammaever work out to be positive, or is it always negative? – Michael Seifert Sep 02 '21 at 18:14