I believe there are some big misunderstandings in the question...
First: usually error terms are additive, not multiplicative.
There are the so-called multiplicative error models (MEM), but I don't think it's the case here, because usually in MEM the multiplicative error is time-dependent (i.e., $\epsilon_{t} \in [0, \infty)$), while in the OP's question the error term is withdrawn from a fixed interval (i.e, $\epsilon \in [0.5, 1.5]$). A good introduction to MEM can be found here.
Second: This said, it seems that we cannot find confidence intervals for this "model" because it is actually possible to find certainty intervals (sorry if this term doesn't exist, maybe I've just created it...).
Consider, for instance, the original model:
$$
f(x)=\dfrac{1}{e^{(x-2)^2}} + \dfrac{1}{e^{(x+2)^2}}\epsilon,
$$
s.t. $\epsilon \in [0.5, 1.5]$.
If we make a small change to the original model it will be easier to understand how to draw the "certainty intervals". Consider the following modification:
$$
f(x)=\dfrac{1}{e^{(x-2)^2}}\epsilon_{1} + \dfrac{1}{e^{(x+2)^2}}\epsilon_{2}.
$$
If we set $\epsilon_{1} \in [1, 1]$ and $\epsilon_{2} \in [0.5, 1.5]$ we have the original model.
Now we can show that the certainty intervals are dependent upon the error terms ($\epsilon_{1}$ and $\epsilon_{2}$) intervals.
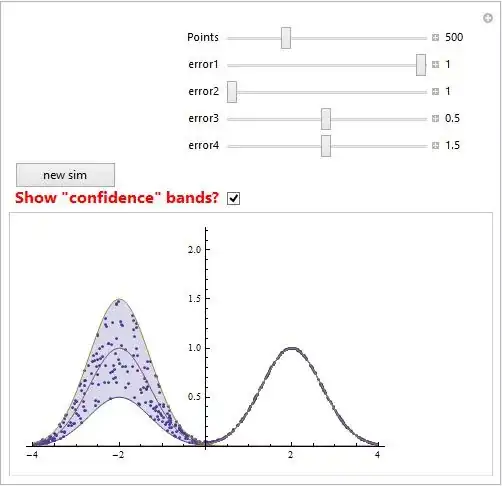
Manipulate[
Module[{f, g, h, y},
{f[y_] := error1*Exp[-(y - 2)^2],
g[y_] := (error1 + error2)/2*Exp[-(y - 2)^2],
h[y_] := error2*Exp[-(y - 2)^2],
G1 = Plot[{f[y], g[y], h[y]}, {y, -4, 4}, Filling -> {1 -> {3}},
PlotRange -> {{-4, 4}, {0, 2}}]}];
Module[{f, g, h, y}, {f[y_] := error3*Exp[-(y + 2)^2],
g[y_] := (error3 + error4)/2*Exp[-(y + 2)^2],
h[y_] := error4*Exp[-(y + 2)^2],
G2 = Plot[{f[y], g[y], h[y]}, {y, -4, 4}, Filling -> {1 -> {3}},
PlotRange -> {{-4, 4}, {0, 2}}]}];
sim = Table[{x,Exp[-(x - 2)^2]*RandomReal[{error1, error2}] +
Exp[-(x + 2)^2]*RandomReal[{error3, error4}]},
{x,RandomReal[{-4, 4}, Points]}];
G3 = ListPlot[sim, PlotStyle -> Thick];
Show[If[bands, {G3, G2, G1}, G3], PlotRange -> {{-4, 4}, {0, 2.2}}],
{{Points, 500}, 100, 1500, Appearance -> "Labeled"},
{{error1, 1}, 0, 1, Appearance -> "Labeled"},
{{error2, 1}, 1, 2, Appearance -> "Labeled"},
{{error3, 0.5}, 0, 1, Appearance -> "Labeled"},
{{error4, 1.5}, 1, 2, Appearance -> "Labeled"},
Button["new sim", {Clear@sim, sim}, ImageSize -> 100],
{{bands, False, Style["Show \"confidence\" bands?", Bold, Red,
FontSize -> 16]}, {True, False}}]




Rand hope someone has contributed a model that might be appropriate for your situation. – whuber May 21 '13 at 15:23