I need to visualize 3D data having 200*200 data points.
ListDensityPlot is a good candidate, but it seems to have strange performance issues.
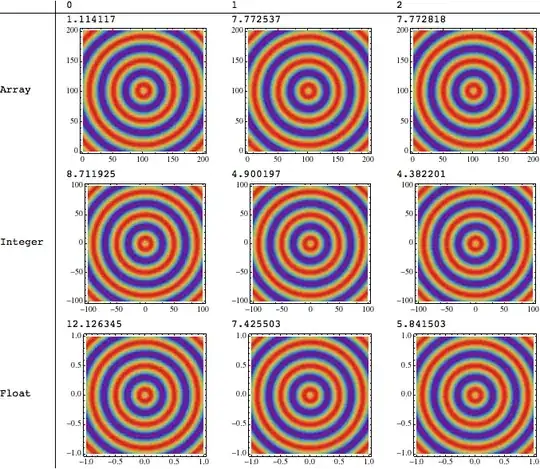
I created a small test that plots 40000 points. It has three cases: using 2D array as input, using array of 3D points with integer coordinates $(x,y)$ and using array of 3D points with float coordinates $(x,y)$.
TestFunction[x_, y_] := Sin[Pi/20* Sqrt[x^2 + y^2]];
testdata = Table[TestFunction[i, j], {i, -100, 100}, {j, -100, 100}];
testdataPoints =
Flatten[Table[{i, j, TestFunction[i, j]}, {i, -100, 100}, {j, -100,
100}], 1];
testdataPoints2 =
Flatten[Table[{i*0.01, j*0.01, TestFunction[i, j]}, {i, -100,
100}, {j, -100, 100}], 1];
Benchmark[d_, n_] :=
Timing[ListDensityPlot[d, ColorFunction -> "Rainbow",
PlotRange -> Full, InterpolationOrder -> n]];
TableForm[
Table[Benchmark[data,
n], {data, {testdata, testdataPoints, testdataPoints2}}, {n, 0,
2}], TableHeadings -> {{"Array", "Integer", "Float"}, {0, 1, 2}}]

It gives pretty strange result. When I simply scale axes and my coordinates are not integer anymore (the case is called "Float"). The performance drops almost 10 times (3 seconds vs 25 seconds).
Here's the output timing:
0 1 2
Array 0.834276 2.96802 3.05685
Integer 4.84968 3.42562 3.19835
Float 27.5574 26.1669 25.9262
Any explanation for such behavior?
Edit
As alternative to Michael's solution one can use ArrayPlot (as Silvia suggested) if interpolation is not important.
It can be easily scaled to look like "Float" case.
Developer`PackedArrayQ[]on your three lists? – J. M.'s missing motivation Jun 18 '13 at 15:15Developer`PackedArrayQ /@ {testdata, testdataPoints, testdataPoints2}gives{False, False, False}. – BlacKow Jun 18 '13 at 15:19Interpolation,DensityPlotandPlotPoints -> 101is faster for me thanListDensityPlot. (Not an answer to your Q, though.) – Michael E2 Jun 18 '13 at 15:20packedtestdata = Developer`ToPackedArray[testdata]). – J. M.'s missing motivation Jun 18 '13 at 15:21Nto the data first:Developer`ToPackedArray[N[testdata]]. – Michael E2 Jun 18 '13 at 15:24ListDensityPlotis very inefficient. See this Q&A for some details. – rm -rf Jun 18 '13 at 15:25packedtestdata = Developer``ToPackedArray /@ {N[testdata], N[testdataPoints], N[testdataPoints2]};Gives same result, maybe a fraction of second less – BlacKow Jun 18 '13 at 15:33ListDensityPlot, that is theArrayPlot. – Silvia Jun 18 '13 at 15:59FrameTickswhich is an additional effort – BlacKow Jun 18 '13 at 16:29