WARNING: a couple of days ago I posted a similar question, but due to the impossibility of DirichletCondition[] to handle "cross-coupling of dependent variables" I thought of reformulating the same problem using a single function.
The goal is to find a function $f : \Omega \subset \mathbb{R}^2 \to \mathbb{R}$ such that:
$$ \begin{cases} \Delta f = 0 & \text{on} \; \Omega \\ \frac{\text{d}}{\text{d} n} f = y\,n_x - x\,n_y & \text{on} \; \partial\Omega \end{cases} $$
with $n = (n_x,\,n_y)$ exterior normal vector to $\partial\Omega$.
Therefore, once the following plane region is defined:
c[x_, y_] := 2 x^2 + 3 y^2
{nx, ny} = Grad[c[x, y], {x, y}];
Ω = ImplicitRegion[c[x, y] <= 1, {x, y}];
RegionPlot[Ω, AspectRatio -> Automatic]
I tried to set the problem by writing:
bcs = NeumannValue[y nx - x ny, True];
fsol = NDSolveValue[Laplacian[f[x, y], {x, y}] == bcs, f, {x, y} ∈ Ω];
Plot3D[fsol[x, y], {x, y} ∈ Ω]
but unfortunately I got tons of warnings:
NDSolveValue::femibcnd: No DirichletCondition or Robin-type NeumannValue was specified for {f}; the result may not be unique.
LinearSolve::parpiv: Zero pivot was detected during the numerical factorization or there was a problem in the iterative refinement process. It is possible that the matrix is ill-conditioned or singular.
NDSolveValue::fempsf: PDESolve could not find a solution.
and I just can't figure out what I'm doing wrong. Ideas? Thank you!
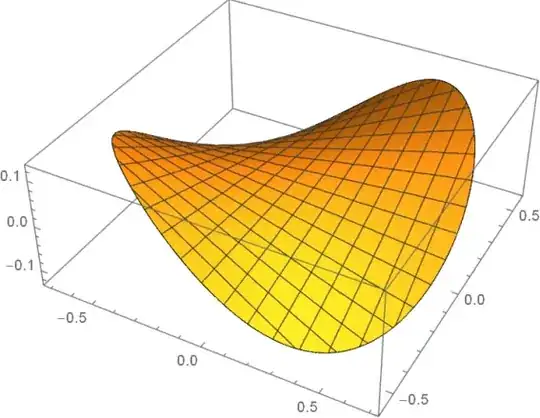
EDIT 1: copying the code of user21 I wrote the following routine:
Needs["NDSolve`FEM`"];
DiniNeumann[f_, Ω_] := Module[
{dpde, dbc, vd, sd, mdata, g, load, stiffness, damping, mass,
prec, dof, loadContribution, stiffnessContribution, initCoeffs,
constraintMatrix, constraintRows, constraintValueMatrix},
{dpde, dbc, vd, sd, mdata} = Quiet[ProcessPDEEquations[
{Laplacian[g[x, y], {x, y}] == NeumannValue[f, True]}, g,
{x, y} ∈ Ω]]; {load, stiffness, damping, mass} = dpde["All"];
DeployBoundaryConditions[{load, stiffness}, dbc];
prec = mdata["Precision"]; dof = mdata["DegreesOfFreedom"];
loadContribution = SparseArray[{}, {dof, 1}, N[0, prec]];
stiffnessContribution = SparseArray[{}, {dof, dof}, N[0, prec]];
initCoeffs = InitializePDECoefficients[vd, sd,
"LoadCoefficients" -> {{1}}];
constraintMatrix = Transpose[DiscretizePDE[initCoeffs, mdata,
sd]["LoadVector"]]; constraintRows = {1};
constraintValueMatrix = SparseArray[{{N[0, prec]}}];
dbc = DiscretizedBoundaryConditionData[{loadContribution,
stiffnessContribution, constraintMatrix, constraintRows,
constraintValueMatrix, {dof, 0, Length[constraintRows]}}, 1];
DeployBoundaryConditions[{load, stiffness}, dbc,
"ConstraintMethod" -> "Append"]; ElementMeshInterpolation[{Ω},
Take[LinearSolve[stiffness, load], mdata["DegreesOfFreedom"]]]]
thanks to which it's sufficient to write:
Ω = ToElementMesh@ImplicitRegion[2 x^2 + 3 y^2 <= 1, {x, y}];
f = DiniNeumann[y (4 x) - x (6 y), Ω];
Plot3D[f[x, y], {x, y} ∈ Ω]
to get what you want:
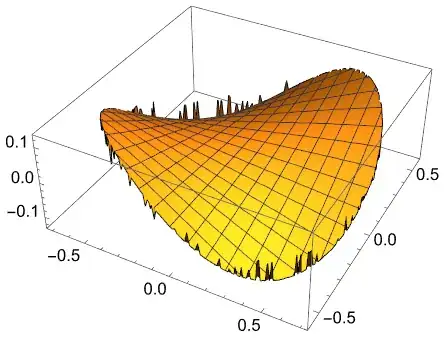
EDIT 2: thanks to suggestions from Ulrich Neumann and user21, writing:
Needs["NDSolve`FEM`"];
Ω = ToElementMesh[ImplicitRegion[2 x^2 + 3 y^2 <= 1, {x, y}],
"IncludePoints" -> {{0, 0}}];
fsol = NDSolveValue[{Laplacian[f[x, y], {x, y}] == NeumannValue[y (4 x) - x (6 y),
True], DirichletCondition[f[x, y] == 0, x == 0 && y == 0]},
f, {x, y} ∈ Ω];
Plot3D[fsol[x, y], {x, y} ∈ Ω]
I get:
which apparently is lower quality than what you got above (at least using the version 13.1.0 for Microsoft Windows (64-bit) (August 14, 2022)).
EDIT 3: in version 13.2.0 for Microsoft Windows (64-bit) (November 18, 2022) the plot is smooth, all fixed!