Using the Geodesics in Heat Algorithm implemented here, we can calculate the distances of all vertices on the surface to a given vertex. By repeating this algorithm on a selected subset of vertices on the surface, we can calculate readily how close all the other vertices on these surfaces are, and label them according to which selected vertex they are closest to.
This can be done with the following code, with the Stanford bunny example again:
a = DiscretizeGraphics[ExampleData[{"Geometry3D", "StanfordBunny"}]]
prep = heatDistprep[a];
npoints = 10;
nvertices = prep[[5]];
vertices = prep[[6]];
faces = MeshCells[a, 2] /. Polygon[p_] :> p;
phiall = {};
randvertlist =
DeleteDuplicates[RandomInteger[{1, nvertices}, npoints]];
npoints = Length[randvertlist];
i = 1;
While[i < npoints + 1,
phi = solveHeat[a, prep, randvertlist[[i]], 0.5];
AppendTo[phiall, phi[[1]]];
i++];
labels = Map[Ordering[phiall[[All, #]]][[1]] &, Range[nvertices]]/npoints;
plotdata =
Map[Join[vertices[[#]], {labels[[#]]}] &, Range[Length[vertices]]];
labelplot =
Graphics3D[{EdgeForm[],
GraphicsComplex[vertices, Map[Polygon, faces],
VertexColors ->
Table[ColorData["BrightBands"][labels[[i]]], {i, 1,
nvertices}]]}, Boxed -> False, Lighting -> "Neutral"];
pointplot =
Graphics3D[{Black, Ball[Map[vertices[[#]] &, randvertlist], 0.003]},
Boxed -> False];
Show[{pointplot, labelplot}]

An issue with this approach is that the boundaries in the visualisation are somewhat "rough", and we don't get directly the edges of the Voronoi cells. Any hints on how to do this would be great.
I hope someone finds this useful.
To make the answer self-contained, the Geodesics code is given here:
heatDistprep[mesh0_] := Module[{a = mesh0, vertices, nvertices, edges, edgelengths, nedges, faces, faceareas, unnormfacenormals, acalc, facesnormals, facecenters, nfaces, oppedgevect, wi1, wi2, wi3, sumAr1, sumAr2, sumAr3, areaar, gradmat1, gradmat2, gradmat3, gradOp, arear2, divMat, divOp, Delta, t1, t2, t3, t4, t5, , Ac, ct, wc, deltacot, vertexcoordtrips, adjMat},
vertices = MeshCoordinates[a]; (*List of vertices*)
edges = MeshCells[a, 1] /. Line[p_] :> p; (*List of edges*)
faces = MeshCells[a, 2] /. Polygon[p_] :> p; (*List of faces*)
nvertices = Length[vertices];
nedges = Length[edges];
nfaces = Length[faces];
adjMat = SparseArray[Join[({#1, #2} -> 1) & @@@ edges, ({#2, #1} -> 1) & @@@edges]]; (*Adjacency Matrix for vertices*)
edgelengths = PropertyValue[{a, 1}, MeshCellMeasure];
faceareas = PropertyValue[{a, 2}, MeshCellMeasure];
vertexcoordtrips = Map[vertices[[#]] &, faces];
unnormfacenormals = Cross[#3 - #2, #1 - #2] & @@@ vertexcoordtrips;
acalc = (Norm /@ unnormfacenormals)/2;
facesnormals = Normalize /@ unnormfacenormals;
facecenters = Total[{#1, #2, #3}]/3 & @@@ vertexcoordtrips;
oppedgevect = (#1 - #2) & @@@ Partition[#, 2, 1, 3] & /@vertexcoordtrips;
wi1 = -Cross[oppedgevect[[#, 1]], facesnormals[[#]]] & /@Range[nfaces];
wi2 = -Cross[oppedgevect[[#, 2]], facesnormals[[#]]] & /@Range[nfaces];
wi3 = -Cross[oppedgevect[[#, 3]], facesnormals[[#]]] & /@Range[nfaces];
sumAr1 = SparseArray[Join[Map[{#, faces[[#, 1]]} -> wi1[[#, 1]] &, Range[nfaces]],Map[{#, faces[[#, 2]]} -> wi2[[#, 1]] &, Range[nfaces]],Map[{#, faces[[#, 3]]} -> wi3[[#, 1]] &, Range[nfaces]]]];
sumAr2 = SparseArray[Join[Map[{#, faces[[#, 1]]} -> wi1[[#, 2]] &, Range[nfaces]], Map[{#, faces[[#, 2]]} -> wi2[[#, 2]] &, Range[nfaces]],Map[{#, faces[[#, 3]]} -> wi3[[#, 2]] &, Range[nfaces]]]];
sumAr3 =SparseArray[Join[Map[{#, faces[[#, 1]]} -> wi1[[#, 3]] &, Range[nfaces]], Map[{#, faces[[#, 2]]} -> wi2[[#, 3]] &, Range[nfaces]], Map[{#, faces[[#, 3]]} -> wi3[[#, 3]] &, Range[nfaces]]]];
areaar = SparseArray[Table[{i, i} -> 1/(2*acalc[[i]]), {i, nfaces}]];
gradmat1 = areaar.sumAr1;
gradmat2 = areaar.sumAr2;
gradmat3 = areaar.sumAr3;
gradOp[u_] := Transpose[{gradmat1.u, gradmat2.u, gradmat3.u}];
arear2 = SparseArray[Table[{i, i} -> (2*faceareas[[i]]), {i, nfaces}]];
divMat = {Transpose[gradmat1].arear2, Transpose[gradmat2].arear2,Transpose[gradmat3].arear2};
divOp[q_] := divMat[[1]].q[[All, 1]] + divMat[[2]].q[[All, 2]] + divMat[[3]].q[[All, 3]];
Delta = divMat[[1]].gradmat1 + divMat[[2]].gradmat2 + divMat[[3]].gradmat3;
SetSystemOptions["SparseArrayOptions" -> {"TreatRepeatedEntries" -> 1}]; (*Required to allow addition of value assignment to Sparse Array*)
t1 = Join[faces[[All, 1]], faces[[All, 2]], faces[[All, 3]]];
t2 = Join[acalc, acalc, acalc];
Ac = SparseArray[Table[{t1[[i]], t1[[i]]} -> t2[[i]], {i, nfaces*3}]];
SetSystemOptions["SparseArrayOptions" -> {"TreatRepeatedEntries" -> 0}];
{Ac, Delta, gradOp, divOp, nvertices, vertices, adjMat}
]
solveHeat[mesh0_, prepvals_, i0_, t0_] := Module[{nvertices, delta, t, u, Ac, Delta, g, h, phi, gradOp, divOp, vertices, plotdata},
vertices = prepvals[[6]];
nvertices = prepvals[[5]];
Ac = prepvals[[1]];
Delta = prepvals[[2]];
gradOp = prepvals[[3]];
divOp = prepvals[[4]];
delta = Table[If[i == i0, 1, 0], {i, nvertices}];
t = t0;
u = LinearSolve[(Ac + t*Delta), delta];
g = gradOp[u];
h = -Normalize /@ g;
phi = LinearSolve[Delta, divOp[h]];
plotdata = Map[Join[vertices[[#]], {phi[[#]]}] &, Range[Length[vertices]]];
{phi}
]

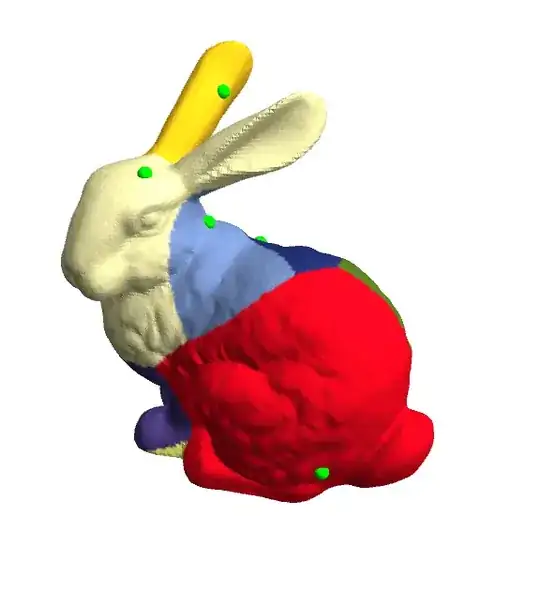
SparseArray[]stuff definitely isn't compilable, and in this case, they're the least memory-intensive storage format. The fuzziness in this case is expected, since at the moment, one is only classifying mesh vertices according to which feature point is nearest. For a sharp boundary, one would need to be able to draw across mesh polygons that might get split. – J. M.'s missing motivation Apr 05 '17 at 22:11